1. Giamarellou H, Poulakou G. Multidrug-resistant gram-negative infections: what are the treatment options? Drugs. 2009; 69:1879–1901. PMID:
19747006.
2. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism
MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016; 16:161–168. PMID:
26603172.
3. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (
mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016; 21:30155. PMID:
26967914.
4. Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, et al.
mcr-1.2, a new
mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing
Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother. 2016; 60:5612–5615. PMID:
27401575.
5. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016; 21:30280.
6. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene
mcr-3 in
Escherichia coli. MBio. 2017; 8:e00543-17. PMID:
28655818.
7. Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid-mediated colistin resistance
mcr-4 gene in
Salmonella and
Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017; 22:30589. PMID:
28797329.
8. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of
mcr-1 in
Escherichia coli from food-producing animals. Lancet Infect Dis. 2016; 16:293. PMID:
26973308.
9. von Wintersdorff CJ, Wolffs PF, van Niekerk JM, Beuken E, van Alphen LB, Stobberingh EE, et al. Detection of the plasmid-mediated colistin-resistance gene
mcr-1 in faecal metagenomes of Dutch travellers. J Antimicrob Chemother. 2016; 71:3416–3419. PMID:
27559117.
10. Robinson TP, Bu DP, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. 2016; 110:377–380. PMID:
27475987.
11. Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, et al. Dissemination and mechanism for the
MCR-1 colistin resistance. PLoS Pathog. 2016; 12:e1005957. PMID:
27893854.
12. Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, et al. Colistin resistance gene
mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 2016; 16:282–283. PMID:
26774242.
13. Snesrud E, Ong AC, Corey B, Kwak YI, Clifford R, Gleeson T, et al. Analysis of Serial Isolates of
mcr-1-positive
Escherichia coli reveals a highly active IS
Apl1 transposon. Antimicrob Agents Chemother. 2017; 61:e00056-17. PMID:
28223389.
14. Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the
mcr-1 colistin resistance gene in carbapenem-resistant
Enterobacteriaceae. Lancet Infect Dis. 2016; 16:287–288. PMID:
26842776.
15. Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, et al. Possible transmission of
mcr-1-harboring
Escherichia coli between companion animals and human. Emerg Infect Dis. 2016; 22:1679–1681. PMID:
27191649.
16. Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, et al. Emergence of NDM-5- and
MCR-1-producing
Escherichia coli clones ST648 and ST156 from a single muscovy duck (
Cairina moschata). Antimicrob Agents Chemother. 2016; 60:6899–6902. PMID:
27550364.
17. Teo JQ, Ong RT, Xia E, Koh TH, Khor CC, Lee SJ, et al.
mcr-1 in multidrug-resistant
blaKPC-2-producing clinical
Enterobacteriaceae isolates in Singapore. Antimicrob Agents Chemother. 2016; 60:6435–6437. PMID:
27503652.
18. Sun J, Li XP, Yang RS, Fang LX, Huo W, Li SM, et al. Complete nucleotide sequence of an IncI2 plasmid coharboring
blaCTX-M-55 and
mcr-1. Antimicrob Agents Chemother. 2016; 60:5014–5017. PMID:
27216063.
19. Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, et al. Prevalence of
mcr-1 in
Escherichia coli and
Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017; 17:400–410. PMID:
28139430.
20. Moellering RC Jr. NDM-1—a cause for worldwide concern. N Engl J Med. 2010; 363:2377–2379. PMID:
21158655.
21. Lim SK, Kang HY, Lee K, Moon DC, Lee HS, Jung SC. First detection of the
mcr-1 gene in
Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob Agents Chemother. 2016; 60:6991–6993. PMID:
27572390.
23. Hong JS, Yoon EJ, Lee H, Jeong SH, Lee K. Clonal Dissemination of
Pseudomonas aeruginosa sequence type 235 isolates carrying
blaIMP-6 and emergence of
blaGES-24 and
blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother. 2016; 60:7216–7223. PMID:
27671068.
24. Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of
MCR-1-producing colistin-resistant
Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016; 60:4394–4397. PMID:
27161623.
25. Kim ES, Chong YP, Park SJ, Kim MN, Kim SH, Lee SO, et al. Detection and genetic features of
MCR-1-producing plasmid in human
Escherichia coli infection in South Korea. Diagn Microbiol Infect Dis. 2017; 89:158–160. PMID:
28780246.
26. Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, et al. First report of the globally disseminated IncX4 plasmid carrying the
mcr-1 gene in a colistin-resistant
Escherichia coli Sequence Type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother. 2016; 60:6415–6417. PMID:
27503650.
27. Elnahriry SS, Khalifa HO, Soliman AM, Ahmed AM, Hussein AM, Shimamoto T, et al. Emergence of plasmid-mediated colistin resistance gene
mcr-1 in a clinical
Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016; 60:3249–3250. PMID:
26953204.
28. Bradley DE, Coetzee JN. The determination of two morphologically distinct types of pilus by plasmids of incompatibility group I2. J Gen Microbiol. 1982; 128:1923–1926. PMID:
6128374.
29. Yoshida T, Kim SR, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999; 181:2038–2043. PMID:
10094679.
30. Akahane K, Sakai D, Furuya N, Komano T. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol Genet Genomics. 2005; 273:350–359. PMID:
15838638.
31. Komano T, Kim SR, Yoshida T, Nisioka T. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994; 243:6–9. PMID:
7932741.
32. Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, et al. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the
mcr-1 acquisition. Sci Rep. 2017; 7:424. PMID:
28336940.
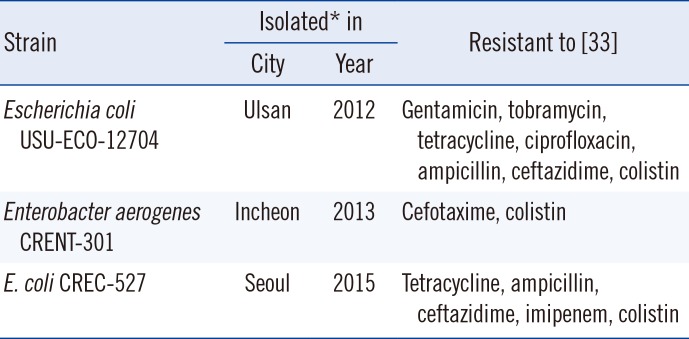
33. Yoon EJ, Yang JW, Kim JO, Lee H, Lee KJ, Jeong SH. Carbapenemase-producing
Enterobacteriaceae in South Korea: a report from the National Laboratory Surveillance System. Future Microbiol. 2018; 13:771–783. PMID:
29478336.
34. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. PMID:
15034147.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download