Abstract
Background
Molecular genetic abnormalities are observed in over 90% of chronic myelomonocytic leukemia (CMML) cases. Recently, several studies have demonstrated the negative prognostic impact of ASXL1 mutations in CMML patients. We evaluated the prognostic impact of ASXL1 mutations and compared five CMML prognostic models in Korean patients with CMML.
Methods
We analyzed data from 36 of 57 patients diagnosed as having CMML from January 2000 to March 2016. ASXL1 mutation analysis was performed by direct sequencing, and the clinical and laboratory features of patients were compared according to ASXL1 mutation status.
Results
ASXL1 mutations were detected in 18 patients (50%). There were no significant differences between the clinical and laboratory characteristics of ASXL1-mutated (ASXL1+) CMML and ASXL1-nonmutated (ASXL1−) CMML patients (all P>0.05). During the median follow-up of 14 months (range, 0–111 months), the overall survival (OS) of ASXL1+ CMML patients was significantly inferior to that of ASXL1− CMML patients with a median survival of 11 months and 19 months, respectively (log-rank P=0.049). An evaluation of OS according to the prognostic models demonstrated inferior survival in patients with a higher risk category according to the Mayo molecular model (log-rank P=0.001); the other scoring systems did not demonstrate a significant association with survival.
Chronic myelomonocytic leukemia (CMML) is a clonal hematologic neoplasm characterized by overlapping features of myelodysplastic and myeloproliferative syndrome with peripheral blood monocytosis [12]. According to the 2016 WHO classification, CMML is further divided into three groups based on peripheral blood (PB) and bone marrow (BM) blast counts: CMML-0 (PB blasts <2%, BM blasts <5%), CMML-1 (PB blasts 2–4%, BM blasts 5–9%), and CMML-2 (PB blasts 5–19%, BM blasts 10–19%) [2]. In addition, distinction according to the French-American-British (FAB) subtypes — proliferative type (white blood cell [WBC] count ≥13×109/L) and dysplastic type (WBC count <13×109/L) — is also recommended [2].
Clonal cytogenetic abnormalities are seen in approximately 20–30% of CMML cases, and molecular genetic abnormalities are seen in over 90% of CMML cases [3456]. The most commonly mutated genes in CMML involve epigenetic regulation (TET2, ~60%), spliceosome machinery (SRSF2, ~50%), chromatin/histone modulation (ASXL1, ~50%), and cell signaling (RAS, ~30%), and these genetic changes can be used as clonal evidence supporting the diagnosis of CMML, particularly in patients with a normal karyotype [2]. With regard to prognosis, to date, only frameshift or nonsense ASXL1 mutations have been consistently reported as a poor prognostic factor, which has led to the incorporation of ASXL1 mutations into prognostic models of CMML [789]. The most common ASXL1 mutation is c.1934dupG (p.Gly646Trpfs*12), which accounts for 40–50% of ASXL1 mutations.
Several prognostic models have been proposed for CMML: the Spanish cytogenetic risk stratification [3], the MD Anderson prognostic scoring system (MDAPS) [10], the CMML-specific prognostic scoring system (CPSS) [11], the Groupe Français des Myélodysplasies (GFM) model [8], the Mayo prognostic model [12], and the Mayo molecular model (MMM) [9]. These models never been validated for Korean patients. The objectives of this study were to evaluate the prognostic impact of ASXL1 mutations and to compare previously reported CMML prognostic models in Korean patients with CMML.
A total of 57 patients were diagnosed as having CMML from January 2000 to March 2016 at Samsung Medical Center, Seoul, Korea. Of them, 21 patients whose samples were unavailable for genetic analysis were excluded, and the remaining 36 were included in this retrospective study. CMML was diagnosed and divided into subtypes according to the 2008 and 2016 WHO classifications [213]. The clinical and laboratory information of patients, including age, sex, complete blood counts, BM blasts (%), cytogenetic study results, and treatment history was collected from electronic medical records (Table 1 and Supplemental Data Table S1). The patients were stratified into different risk categories according to the five prognostic scoring systems: the Spanish cytogenetic risk stratification, MDAPS, the GFM model, the Mayo model, and MMM. The Spanish cytogenetic risk stratification divides patients into low risk (normal karyotype or loss of Y chromosome), high risk (presence of trisomy 8, or abnormalities of chromosome 7, or a complex karyotype), and intermediate risk (all others) [3]. MDAPS stratifies patients into four risk groups based on the following risk factors: hemoglobin <12 g/dL, presence of circulating immature myeloid cells (IMC), absolute lymphocyte count >2.5×109/L, and BM blasts >10% [10]. The GFM model stratifies patients into three risk groups based on age >65 years, WBC count >15×109/L, anemia (<11 g/dL for males and <10 g/dL for females), platelet count <100×109/L, and ASXL1 mutation [8]. The Mayo model stratifies patients into three groups based on the following risk factors: absolute monocyte count (AMC) >10×109/L, presence of circulating IMC, hemoglobin <10 g/dL, and platelet count <100×109/L. MMM has added ASXL1 mutation to the Mayo prognostic model components as a risk factor and stratifies patients into four groups: low (no risk factor), intermediate-1 (one risk factor), intermediate-2 (two risk factors), and high risk (≥3 risk factors) [912]. The study protocol was approved by the Institutional Review Board of Samsung Medical Center, and written informed consent was obtained from all patients.
Genomic DNA was isolated from BM aspirate samples at the time of initial diagnosis using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA), according to the manufacturer's instructions, and ASXL1 mutation analysis was performed on the archived samples. Exon 14 of ASXL1 and its flanking intronic regions were amplified by PCR using primers designed by the authors (See Supplemental Data Table S2) on a Thermal Cycler 9700 (Applied Biosystems, Foster City, CA, USA), with the following cycling conditions: 94℃ for 5 minutes, followed by 32 cycles at 94℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 30 seconds, and a final extension at 72℃ for 7 minutes. Direct sequencing was performed using the ABI Prism 3130xl genetic analyzer (Applied Biosystems) with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems).
Patient characteristics according to ASXL1 mutation status were compared using the chi-square test or Fishe's exact test for categorical variables and the Mann-Whitney test or two sample t-test for continuous variables. Overall survival (OS) was determined from the time of initial diagnosis to death from any cause or last follow-up. Leukemic transformation-free survival (LFS) was determined from the time of initial diagnosis to leukemic transformation, death, or last follow-up. Survival analyses were performed using Kaplan-Meier plots, and differences in survival were compared using the log-rank test. Univariate and multivariate analyses were performed using Cox proportional hazards regression. P<0.05 was considered statistically significant. All statistical analyses were performed using IBM SPSS Statistics version 23 (IBM Corp., Armonk, NY, USA).
An abnormal karyotype was found in ten (27.8%) patients. Trisomy 8 (+8), abnormalities of chromosome 7 (monosomy 7 [−7] and deletion 7q [del7q]), and loss of Y chromosome were each observed in two patients (5.6%) and a complex karyotype including abnormalities of chromosome 3, 5, 6, and 15 was found in one patient (2.8%). ASXL1 mutations were detected in 18 patients (50%). All identified ASXL1 mutations were either frameshift or nonsense mutations, of which four were listed in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic) (Table 2). The most common mutation was c.1934dupG (p.Gly646Trpfs*12), which was found in 12 patients (66.7%). Six unique mutations were detected in the other six patients, of which three were novel: c.1766_1767insAGTA (p.Thr590Valfs*30), c.2973_2985del (p.Leu992Valfs*28), and c.3001dupA (p.Thr1001Asnfs*4). ASXL1 mutations were detected in 14 (54%) patients with a normal karyotype and in four (40%) patients with cytogenetic abnormalities.
Clinical and laboratory characteristics did not significantly differ between ASXL1-mutated (ASXL1+) CMML and ASXL1-nonmutated (ASXL1−) CMML patients (Table 1). Of the different risk stratification classifications, only the GFM prognostic risk categories were significantly different between ASXL1+ CMML and ASXL1− CMML (P=0.006; Table 3).
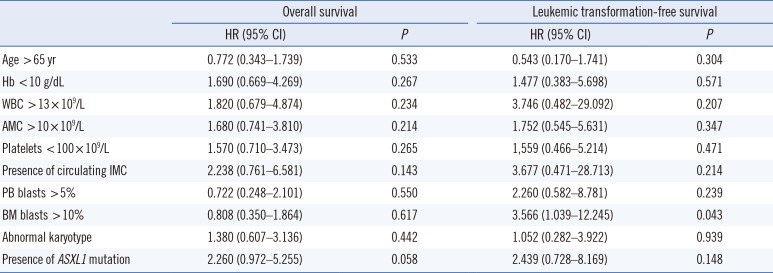
At the median follow-up of 14 months (range, 0–111 months), 33% of the patients had developed leukemic transformation and 72.2% of the patients had expired (median survival 15 months and median LFS 23 months). Leukemic transformation and death occurred more frequently in ASXL1+ CMML than in ASXL1− CMML, but this finding was not statistically significant (50% vs 23.5% and 83.3% vs 61.1%, respectively; Table 1). The OS of ASXL1+ CMML was significantly lower than ASXL1− CMML with a median survival of 11 months and 19 months, respectively (log-rank P=0.049; Fig. 1A). Moreover, the LFS was shorter in ASXL1+ CMML, albeit without statistical significance (log-rank P=0.132; Fig. 1B). Univariate analysis showed that the presence of ASXL1 mutation tended towards inferior survival (Hazard ratio [HR] 2.260, 95% confidence interval [CI] 0.972–5.255; P=0.058) and that BM blasts >10% was associated with inferior LFS (HR 3.566, 95% CI 1.039–12.245; P=0.043; Table 4). However, BM blasts >10% did not retain their prognostic significance in multivariate analysis.
When OS was evaluated according to the prognostic models, survival was significantly inferior in patients with a higher risk category according to MMM, with a median survival of 11 months, 28 months, and 46 months in the high, intermediate-2, and intermediate-1 risk groups, respectively (log-rank P=0.001); however, the other scoring systems did not demonstrate a significant association with survival (all log-rank P>0.05; Fig. 2). According to the Spanish cytogenetic risk stratification, although the survival of the low, intermediate, and high groups was not significantly different (log-rank P=0.064), the survival of the high-risk group was significantly inferior to that of the combined low and intermediate risk groups (log-rank P=0.019).
In our study, ASXL1 mutations were detected in half of the CMML patients, all of which, including three novel mutations, were frameshift or nonsense mutations (Table 2). Approximately 67% of ASXL1 mutations were c.1934dup, and no other mutation hotspots were found. With regard to genotype-phenotype correlations, previous studies have shown an association between ASXL1 mutations and proliferative phenotypes including higher WBC, higher AMC, and the presence of circulating IMC [59]. However, we found no significant difference between the clinical or laboratory characteristics of ASXL1+ and ASXL− CMML patients, which might be due to the limited number of patients. Of note, a higher tendency of WBC and AMC median values was observed in ASXL1+ CMML than in ASXL1− CMML patients. Cytogenetic abnormalities have been reported in approximately 20–30% of CMML patients, and +8, loss of Y, −7/del7q, and trisomy 21 (+21) have been established as common abnormalities [34514]. Our study showed similar findings; approximately 28% of our patients had cytogenetic abnormalities, and 70% of the cytogenetic abnormalities were +8 (20%), loss of Y (20%), −7/del7q (20%), and +21 (10%). Regarding the frequency of ASXL1 mutations, a previous study has demonstrated that ASXL1 mutations cluster with an abnormal karyotype (45% vs 34%, P=0.04) [4]; however, in our series, the difference was not statistically significant (40% vs 54%, P=0.457).
The natural course of CMML is highly variable and the OS ranges from 12 months to 35 months [3891011]. Furthermore, in a recent study involving young adult (≤65 years) CMML patients, the median survival was 55 months [15]. These variable clinical courses led to the development of various risk stratification models based on either complete blood counts or cytogenetic or molecular abnormalities. The Spanish cytogenetic risk stratification categorizes patients into three groups; an initial study reported the 5-year OS in the low, intermediate, and high-risk groups to be 35%, 26%, and 4%, respectively [3]. A study on 417 CMML patients demonstrated a significantly different OS for each risk group, with a median survival of 33 months, 24 months, and 14 months for the low, intermediate, and high-risk groups, respectively [14]. In our study, only the high-risk group with +8, −7/del7q, or a complex karyotype showed significantly inferior survival compared with the low/intermediate risk groups; the median survival was 17 months and 1 month for the low/intermediate and high-risk groups, respectively, which was lower than in a previous study [14].
The inferior survival in ASXL1+ CMML patients was first demonstrated in a study by Gelsi-Boyer et al [7], which included 53 CMML patients. Subsequently, GFM has demonstrated the independent poor prognostic value of ASXL1 mutation in 312 CMML patients and thus incorporated ASXL1 mutation into the GFM prognostic model [8]. In contrast, the initial Mayo Clinic study found no prognostic impact for ASXL1 mutation in 226 CMML patients, which led to the development of the Mayo prognostic model without incorporation of ASXL1 mutation. While only ASXL1 nonsense and frameshift mutations were included in the GFM study, all nucleotide variations, including missense variation, were included as ASXL1 mutations in the Mayo Clinic study, which is thought to account for the conflicting prognostic impact of the ASXL1 mutation between the two studies [9]. To further clarify the prognostic relevance of ASXL1 mutation, 466 CMML patients from the Mayo Clinic and the GFM study were analyzed; only frameshift and nonsense ASXL1 mutations were demonstrated to constitute as an independent prognostic factor for OS, but not for leukemic transformation [9]. Therefore, the new MMM was developed by adding ASXL1 mutation to the Mayo model. We also demonstrated the inferior prognostic impact of ASXL1 mutation in Korean patients with CMML (Fig. 1A). When previous prognostic scoring systems were applied to our patients, the survival rates of the different risk groups were not significantly different according to the risk stratification of MDAPS, the GFM prognostic model, or the Mayo model (Fig. 2B–D). Only MMM stratification showed a significant difference between the risk groups (Fig. 2E), and median survival rates were similar to that of a previous MMM study (the median survival of the low, intermediate-1, intermediate-2, and high-risk groups was 97, 59, 31, and 16 months, respectively) [9]. There were no low risk group patients in our study, presumably because of the limited number of patients. Previously, the MDAPS and MMM were also compared for 146 CMML patients in Spain [16]. In contrast to our results, the OS was significantly different in each of the MDAPS risk groups, and in the Mayo model, the low/intermediate-risk groups exhibited better OS than the high-risk groups, although there were no differences between the low and intermediate-risk groups.
Leukemic transformation is one of the main causes of death in CMML, and its incidence ranges from 15% to 30% [9101112]. While Gelsi-Boyer et al [7] showed that ASXL1 mutation is also associated with leukemic transformation, Patnaik et al [17] reported that the risk factors for leukemic transformation are the presence of circulating blasts and female sex, but not the karyotype or mutational status of ASXL1. In our study, leukemic transformation occurred in 33.3% of patients, more frequently in ASXL1+ CMML patients (50%) than in ASXL1− patients (23.5%), but this finding was not statistically significant. Based on univariate analysis, only BM blasts >10% were associated with the risk of leukemic transformation.
This study had some limitations. Our study did not enroll a sufficiently large number of CMML patients. Moreover, we did not evaluate any other genetic abnormalities (i.e., TET2, DNMT3A). Therefore, further large-scale studies using targeted next-generation sequencing are needed.
In conclusion, ASXL1 mutations found in half of the Korean CMML patients in this study were associated with inferior survival. Of the risk classification models, the MMM best stratified the patient risk groups despite the limited number of patients. ASXL1 mutation status needs to be determined for risk stratification in Korean patients with CMML.
References
1. Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2016 update on diagnosis, risk stratification, and management. Am J Hematol. 2016; 91:631–642. PMID: 27185207.
2. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016; 127:2391–2405. PMID: 27069254.
3. Such E, Cervera J, Costa D, Solé F, Vallespi T, Luño E, et al. Cytogenetic risk stratification in chronic myelomonocytic leukemia. Haematologica. 2011; 96:375–383. PMID: 21109693.
4. Wassie EA, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: a Mayo Clinic-French Consortium Study. Am J Hematol. 2014; 89:1111–1115. PMID: 25195656.
5. Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016; 6:e393. PMID: 26849014.
6. Patnaik MM, Lasho TL, Vijayvargiya P, Finke CM, Hanson CA, Ketterling RP, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016; 6:e385. PMID: 26771811.
7. Gelsi-Boyer V, Trouplin V, Roquain J, Adélaïde J, Carbuccia N, Esterni B, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010; 151:365–375. PMID: 20880116.
8. Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013; 31:2428–2436. PMID: 23690417.
9. Patnaik MM, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014; 28:2206–2212. PMID: 24695057.
10. Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002; 99:840–849. PMID: 11806985.
11. Such E, Germing U, Malcovati L, Cervera J, Kuendgen A, Della Porta MG, et al. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood. 2013; 121:3005–3015. PMID: 23372164.
12. Patnaik MM, Padron E, LaBorde RR, Lasho TL, Finke CM, Hanson CA, et al. Mayo prognostic model for WHO-defined chronic myelomonocytic leukemia: ASXL1 and spliceosome component mutations and outcomes. Leukemia. 2013; 27:1504–1510. PMID: 23531518.
13. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009; 114:937–951. PMID: 19357394.
14. Tang G, Zhang L, Fu B, Hu J, Lu X, Hu S, et al. Cytogenetic risk stratification of 417 patients with chronic myelomonocytic leukemia from a single institution. Am J Hematol. 2014; 89:813–818. PMID: 24782398.
15. Patnaik MM, Wassie EA, Padron E, Onida F, Itzykson R, Lasho TL, et al. Chronic myelomonocytic leukemia in younger patients: molecular and cytogenetic predictors of survival and treatment outcome. Blood Cancer J. 2015; 5:e270. PMID: 25555161.
16. Calvo X, Nomdedeu M, Santacruz R, Martínez N, Costa D, Pereira A, et al. Comparison of three prognostic scoring systems in a series of 146 cases of chronic myelomonocytic leukemia (CMML): MD Anderson prognostic score (MDAPS), CMML-specific prognostic scoring system (CPSS) and Mayo prognostic model. A detailed review of prognostic factors in CMML. Leuk Res. 2015; 39:1146–1153.
17. Patnaik MM, Wassie EA, Lasho TL, Hanson CA, Ketterling R, Tefferi A. Blast transformation in chronic myelomonocytic leukemia: risk factors, genetic features, survival, and treatment outcome. Am J Hematol. 2015; 90:411–416. PMID: 25645147.
SUPPLEMENTARY MATERIALS
Supplemental Data Table S1
Detailed clinical and laboratory features of 36 patients with CMML
Fig. 1
Kaplan-Meier plots of the (A) overall survival and (B) leukemic transformation-free survival in patients with chronic myelomonocytic leukemia according to ASXL1 mutation status.
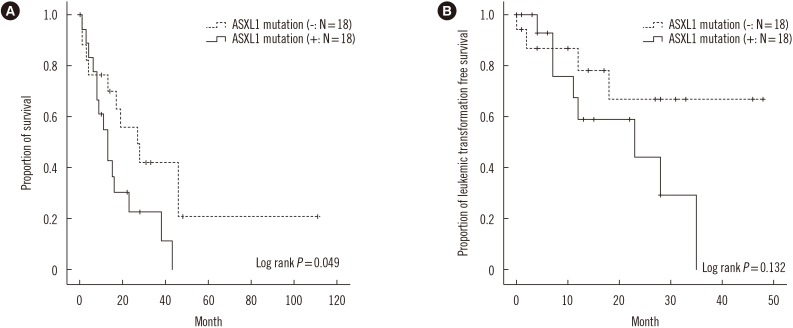
Fig. 2
Kaplan-Meier plots of the overall survival in patients with chronic myelomonocytic leukemia according to the (A) Spanish cytogenetic risk stratification, (B) MDAPS, (C) GFM model, (D) Mayo model, and (E) Mayo molecular model.
Abbreviations: MDAPS, MD Anderson prognostic scoring system; GFM, Groupe Français des Myélodysplasies.
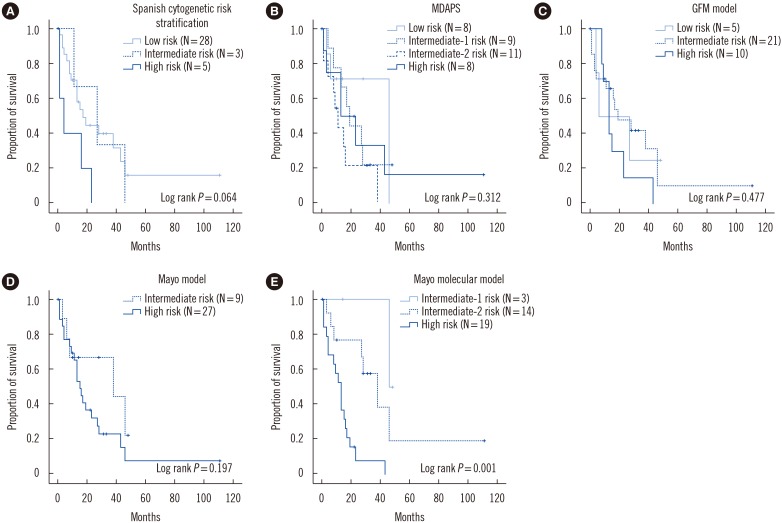
Table 1
Clinical and laboratory characteristics of 36 patients with CMML
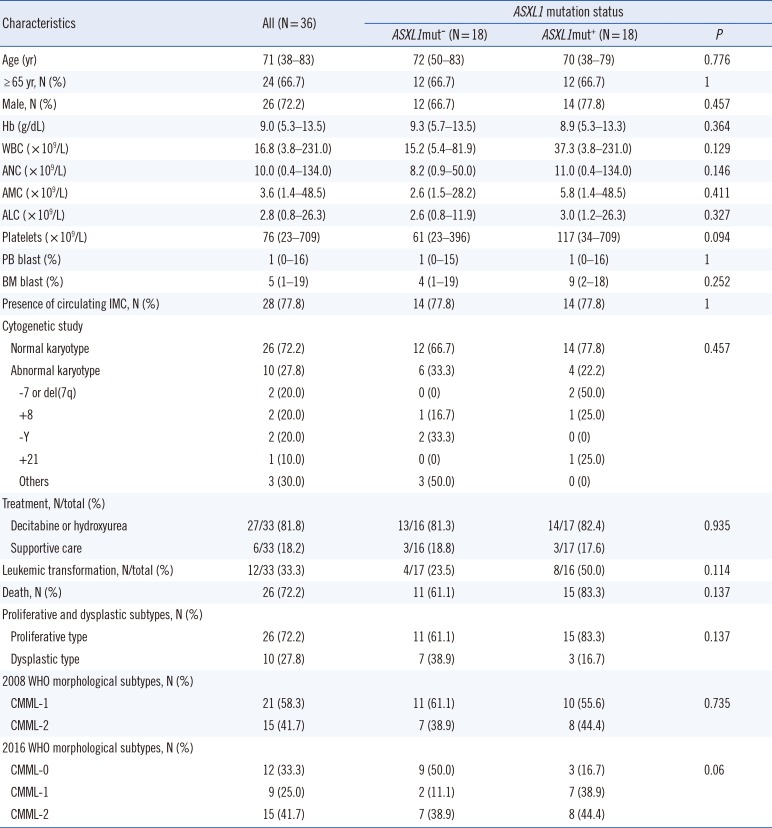
Table 2
Spectrum of ASXL1 mutations in 18 patients with chronic myelomonocytic leukemia
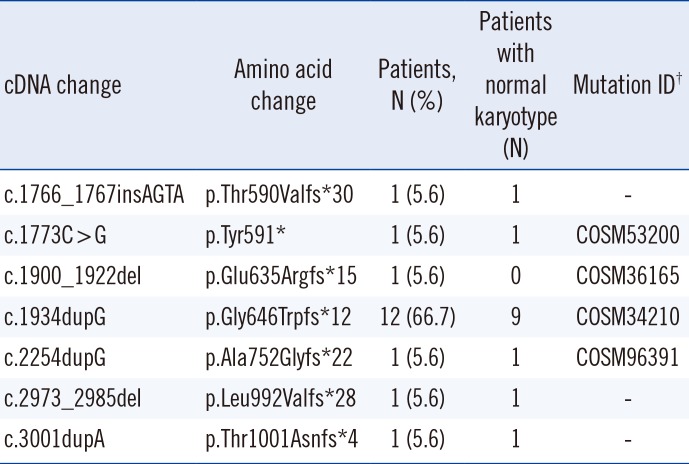
†Mutation ID was assigned based on the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic).
Table 3
Risk stratification of 36 patients with chronic myelomonocytic leukemia according to five different risk stratification models
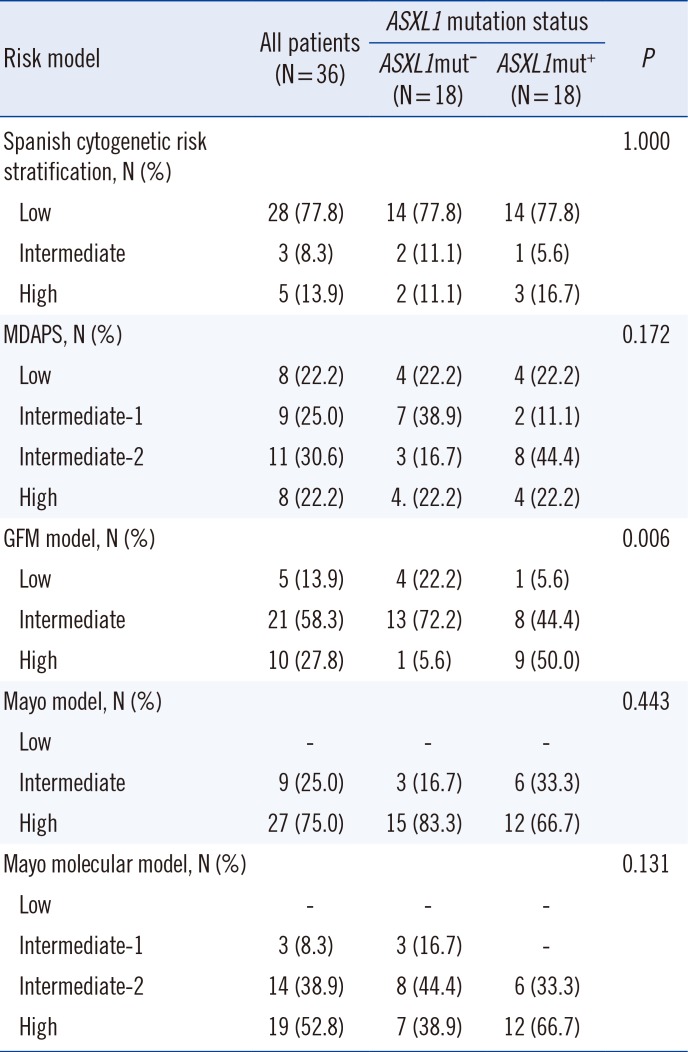
Table 4
Univariate analysis for overall survival and leukemic transformation-free survival in 36 patients with chronic myelomonocytic leukemia




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download