Abstract
Objectives
Blood culture is a one of the most important procedure for diagnosis and treatment of infectious disease, but distribution of pathogenic species and the antimicrobial susceptibility can be vary from pathogen, individual trait, regional or environmental features. In this study, we investigated the changes in frequency of occurrence and antimicrobial susceptibility pattern of blood isolates from 2005 to 2014.
Methods
Data of blood isolates from Kosin Gospel Hospital during 2005 to 2014 were analyzed retrospectively. Blood isolates were cultured for 5 days using BACTEC Plus Aerobic/F and BACTEC lytic/10 Anaerobic/F. Identification and antimicrobial susceptibility test was performed using VITEK 1 system, VITEK 2 XL, PHOENIX 100 and conventional method.
Results
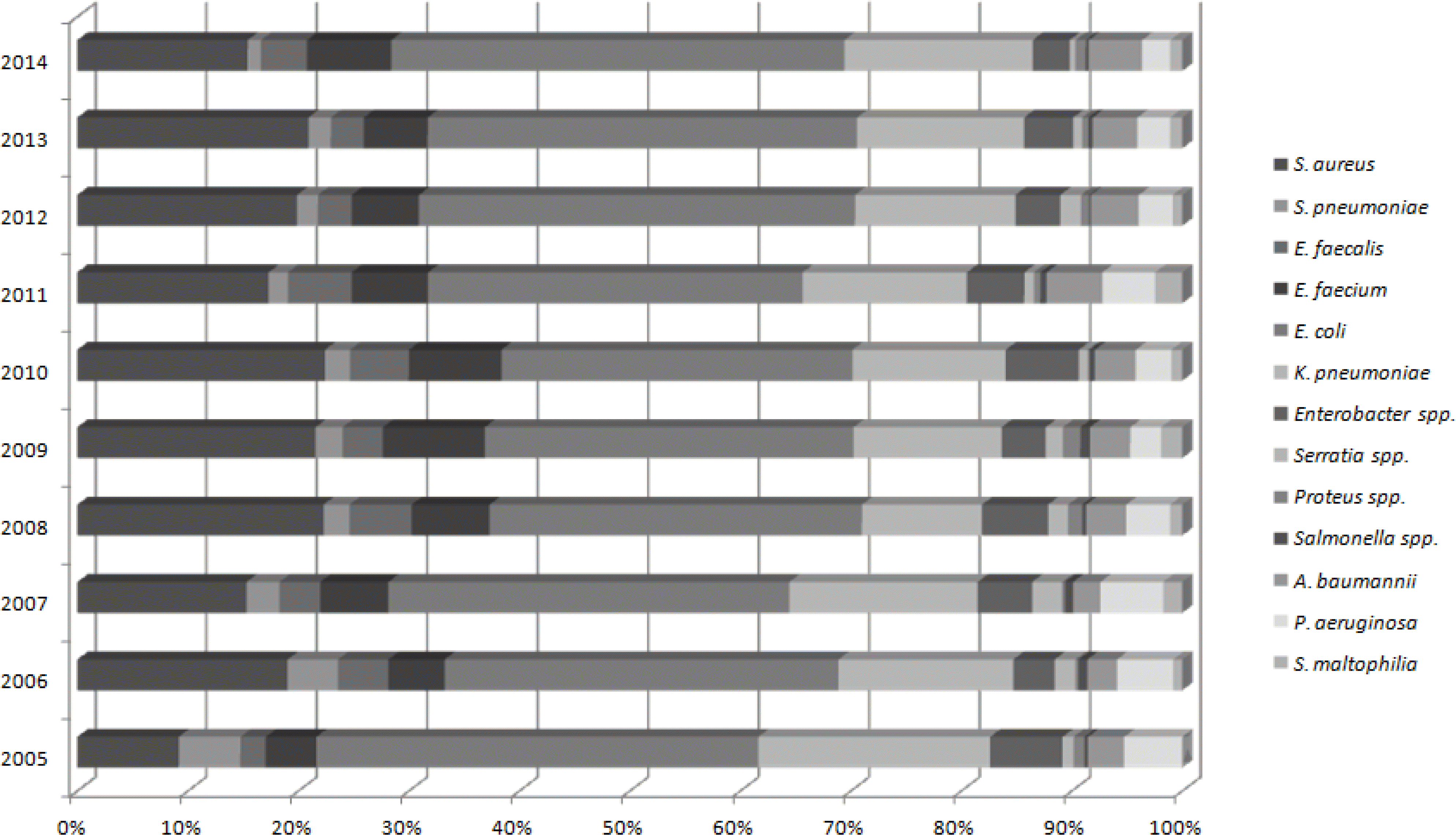
9,847 isolates were identified during 10 years. Among the isolates aerobic or falcutative anaerobic bacteria were isolated in 99.5% specimens, anaerobic were 0.1%, and fugi were 0.4%. Most commonly isolated bacteria were coagulase-negative Staphylococcus (CoNS) followed by Escherichia coli, Staphylococcus aureus and Klebsiella pneumoniae. Candida parapsilosis were most frequently isolated among fungi. The proportion of S. aureus, A. baumannii and E. faecium were increased, while Pseudomonas aeruginosa and Streptococcus pneumoniae decreased over decennium. Imipenem resistant K. pneumoniae were identified. Vancomycin resistant E. faecium and imipenem resistant A. baumannii were increased (7.1% in 2005 to 12.3% in 2014, 0% in 2005 to 55.6% in 2014, respectively).
Go to : 
REFERENCES
1.Washington JA 2nd. Blood cultures: principles and techniques. Mayo Clin Proc. 1975. 50:91–8.
2.Weinstein MP., Reller LB., Murphy JR., Lichtenstein KA. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. I. Laboratory and epidemiologic observations. Rev Infect Dis. 1983. 5:35–53.

3.Arias CA., Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N Engl J Med. 2009. 360:439–43.
4.Kang CI., Song JH. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. 2013. 45:22–31.

5.World Health Organization. Antimicrobial resistance: global report on surveillance. 2014.
6.Bronzwaer S., Lönnroth A., Haigh R. The European Community strategy against antimicrobial resistance. Euro Surveill. 2004. 9:30–4.

7.Murray PR., Baron EJ., Jorgensen JH., Landry ML., Pfaller MA. Manual of clinical microbiology. 9th ed.Washington, DC: American Society for Microbiology Press;2007.
8.Kim NH., Hwang JH., Song KH., Choe PG., Park WB., Kim ES, et al. Changes in Antimicrobial Susceptibility of Blood Isolates in a University Hospital in South Korea, 1998-2010. Infect Chemother. 2012. 44:275–81.

9.Beekmann SE., Diekema DJ., Doern GV. Determining the clinical significance of coagulase-negative staphylococci isolated from blood cultures. Infect Control Hosp Epidemiol. 2005. 26:559–66.

10.Tokars JI. Predictive value of blood cultures positive for coagulase-negative staphylococci: implications for patient care and health care quality assurance. Clin Infect Dis. 2004. 39:333–41.

11.Kim HJ., Lee NY., Kim S., Shin JH., Kim MN., Kim EC, et al. Characteristics of Microorganisms Isolated from Blood Cultures at Nine University Hospitals in Korea during 2009. Korean J Clin Microbiol. 2011. 14:48–54.

12.Kim SY., Lim G., Kim MJ., Suh JT., Lee HJ. Trends in Five-year Blood Cultures of Patients at a University Hospital (2003~2007). Korean J Clin Microbiol. 2009. 12:163–8.

13.Koh EM., Lee SG., Kim CK., Kim M., Yong D., Lee K, et al. Microorganisms isolated from blood cultures and their antimicrobial susceptibility patterns at a university hospital during 1994-2003. Korean J Lab Med. 2007. 27:265–75.

14.Hall KK., Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006. 19:788–802.

15.Yang YL., Hsieh LY., Wang AH., Lo HJ. TSARY Hospitals. Characterization of Candida Species from Different Populations in Taiwan. Mycopathologia. 2011. 172:131–9.
16.Yapar N. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag. 2014. 10:95–105.

17.Trofa D., Gácser A., Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008. 21:606–25.
18.Kim YC., Kim MH., Song JE., Ahn JY., Oh DH., Kweon OM, et al. Trend of methicillin-resistant Staphylococcus aureus (MRSA) bacteremia in an institution with a high rate of MRSA after the reinforcement of antibiotic stewardship and hand hygiene. Am J Infect Control. 2013. 41:e39–43.

19.Gupta N., Limbago BM., Patel JB., Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011. 53:60–7.

20.Hidron AI., Edwards JR., Patel J., Horan TC., Sievert DM., Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008. 29:996–1011.
21.Sanchez GV., Master RN., Clark RB., Fyyaz M., Duvvuri P., Ekta G, et al. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998-2010. Emerg Infect Dis. 2013. 19:133–6.
22.Schwaber MJ., Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J Antimicrob Chemother. 2007. 60:913–20.
Go to : 
 | Fig. 2.Trends in antimicrobial resistance (%) of (A) S. aureus, (B) E. faecalis, (C) E. faecium, (D) E. coli (E) K. pneumonia (F) P. aeruginosa and (G) A. baumannii by year. |
Table 1.
Microorganisms isolated by year
Table 2.
Distribution of blood isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download