Case Report
In September 2014, a 57-year-old man visited the emergency department of National Police Hospital in Seoul, Korea. His chief complaint was dyspnea that had started two days prior. His associated symptoms were cough, sputum, left shoulder pain, diaphoresis, myalgia, fever, and a chilled sensation. He had well-controlled diabetes mellitus which had been diagnosed three years prior. Neither history of admission nor any intervention existed. He was a 30-pack year smoker and had been drinking an average of 100 mg of alcohol twice a week. He had no recent history of loss of consciousness or vomiting. Though he was not a heavy alcoholic, a chance of aspiration remained because he might not have remembered what happened when he was drunk. He usually worked with marble blocks as a mason, with considerable exposure to marble dust in his workplace. The patient insisted that he was always protected by an industrial mask. He reported no other medication, illicit drug, or herbal medicine use except for diabetes medication (linagliptin 2.5 mg and metformin 500 mg, once daily), which he had used ever since his diagnosis. He had no history of sexually transmitted diseases or recent sick contacts. He had traveled to a beach in Incheon one week prior and reported eating raw, unboned gizzards.
At the emergency department, the patient’s initial blood pressure was 206/109 and his heart rate was 123 beats/minute. The blood pressure stabilized spontaneously within an hour, but the sinus tachycardia persisted. His body temperature was 38.2°C. A chest radiograph revealed left hydropneumothorax and pleural adhesion (
Fig. 1). Low-dose chest computed tomography (CT) showed a large amount of left hydropneumothorax, multiloculated left pleural effusion, pleural adhesion, and passive atelectasis of the left lung (
Fig. 2A). In enhanced images, pleural enhancement was observed (
Fig. 2B). Laboratory tests revealed leukocytosis (white blood cell count: 24,300 cells/mm
3) with 93 percent neutrophils. The erythrocyte sedimentation rate and C-reactive protein level were 121 mm/hr and 45.8 mg/dL, respectively. The arterial partial pressure of oxygen was 50.9 mmHg and three liters of oxygen were immediately applied by nasal prong. The hemoglobin A1c level was 6.9%. Aspartate aminotransferase, alanine aminotransferase, and total bilirubin levels were 21 IU/L, 26 IU/L, and 0.5 mg/dL, respectively. The prothrombin time was 1.18 INR and his albumin concentration was 3.0 g/dL, which was consistent with a septic condition. No other significant abnormality was seen in the chemical examinations. Two sets of blood cultures of samples drawn from two different peripheral sites were collected. A pleural fluid sample collected by thoracentesis was turbid, thick, and brownish-yellow in color. It also had a foul odor. The laboratory findings of the sample indicated exudative properties; the pleural fluid lactate dehydrogenase/serum lactate dehydrogenase was 1.08, glucose was 0 mg/dL, his white blood cell count was 2,976 cells/mm
3 with uncountable segmented neutrophils, and the pH was 6.34. One set was cultured using the pleural fluid sample. On hospital day 1, intravenous ceftriaxone, levofloxacin, and clindamycin were administered under the suspicion of parapneumonic empyema due to aspiration pneumonia. A 24-French chest tube was inserted and the patient was admitted to the intensive care unit for close observation. On hospital day 2, to rule out esophageal injury-induced infection, an esophago-gastro-duodenoscopy (EGD) was performed because the patient had a history of eating unboned raw gizzard. The EGD showed no abnormalities, but the possibility of micro-rupture remained.
Figure 1
Chest radiograph on hospital day 1 showing left hydropneumothorax and pleural adhesion.
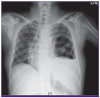
Figure 2
(A) Low-dose chest CT image on hospital day 1 showing a large amount of left hydropneumothorax, multiloculated left pleural effusion, pleural adhesion in left thorax, and passive atelectasis of the left lung. (B) Enhanced chest CT image on hospital day 1 showing a large amount of multiloculated left pleural effusion with pleural enhancement and left pneumothorax, with passive atelectasis with bronchial mucus plugging in the left lung.

Three sets of culture were incubated for 24-48 hours and then inoculated to blood agar and MacConkey plates. The cultured microorganisms were gram-positive cocci, catalase-positive, and coagulase-negative. Colonies were collected from the plates for identification using an automated system (MicroScan WalkAway 96 SI system, Siemens Healthcare Diagnostics Inc., West Sacramento, CA, USA, using conventional Pos panel). On hospital day 6, from all three culture sets, identical organism types were revealed, which were identified as K. kristinae. The biochemical results were negative for crystal violet, micrococcus screen, nitrate, novobiocin, indoxyl phosphatase, Voges-Proskauer, phosphatase, 40% bile esculin, L-Pyrrolidonyl-beta- naphthylamide, p-nitrophenyl-beta-D-galactopyranoside, urea, mannitol, arabinose, bacitracin, inulin, sodium chloride 6.5%, pyruvate, raffinose, ribose, and sorbitol sections of the panel. The optochin, arginine, lactose, trehalose, and mannose sections showed positive results. The overall probability of the identification was 95%.
According to the probability table of the MicroScan WalkAway 96 SI system, it was very rare for K. kristinae to show a negative reaction to the optochin and mannose parts of the panel, and uncommon to show a negative reaction to the trehalose part of the panel. A positive optochin reaction meant that the organism successfully grew in optochin, while positive results in the mannose and trehalose sections meant that the organism could ferment the corresponding carbohydrates and form acid metabolites. Conversely, it was very rare for K. kristinae to react positively to the crystal violet, novobiocin, nitrate, p-nitrophenyl-beta-D-glucuronide, phosphatase, and mannitol sections. This meant that the organism could not survive in low concentrations of crystal violet and novobiocin, could not reduce nitrate to nitrite, could not catalyze the hydrolyzation of p-nitrophenyl-beta-D-galactopyranoside, did not produce alkaline phosphatase, and could not ferment mannitol, respectively. Other sections of the panel may show variable results (MicroScan WalkAway 96 SI system, Siemens Healthcare Diagnostics Inc., USA).
On hospital day 8, the patient was referred to the general ward, as his general condition and laboratory results had improved. On hospital day 11, his antibiotic treatment was escalated to intravenous piperacillin-tazobactam and levofloxacin due to a short episode of relapsed fever (up to 37.8°C). On hospital day 23, since the patient presented a fine general condition and improved chest radiographic findings without any respiratory symptoms or any other laboratory abnormalities, he was discharged with amoxicillin-clavulanic acid prescription for seven days. A follow-up blood culture sample drawn on hospital day 10 was negative and the chest radiographic findings had improved dramatically by the day of discharge.
Discussion
After the taxonomic dissection of Kocuria from the Micrococcus genus, a number of cases of infection have been reported in which these bacteria have been emphasized as emerging pathogens in immunocompromised hosts and hospital-associated infections.
This report described the first case of K. kristinae-induced bacteremic empyema in a healthy man without any history of hospitalization. The only medical history he had was diabetes, which was diagnosed only three years before the event, and was well-controlled, with a hemoglobin A1c level of 6.9% at the time of his admission.
The route of infection was not clear. The first possibility was aspiration pneumonia-induced empyema. Though the patient denied recent vomiting or losing consciousness, there still was a possibility of him lying or just not remembering the event after drinking. The second possibility that we considered was micro-perforation of the esophagus due to raw fish bones. Though we could not find visible esophageal mucosal defects in the EGD, there was a chance that we could not observe micro-injury of the esophagus. A micro-perforation of the esophagus might have acted as an inoculation route of bacteria into the intrathoracic area. The empyemic fluid from the nearby esophagus (
Fig. 3) supported this second idea.
Figure 3
Enhanced chest CT image on hospital day 1 showing effusion near the mediastinum. Nearby esophagus(A) and pericardium (B).

In this case, we started empirical antibiotic therapy to cover the common pathogens of aspiration pneumonia and empyema. Fortunately, the majority of
Kocuria species are, so far, sensitive to most of the antibiotics used empirically in community-acquired pneumonia [
12]. Therefore, regardless of the possibility of infection caused by
K. kristinae, it seemed safer to start empirical antibiotics based on the epidemiology, infection site, and condition of the patient.
We reviewed the literature and identified a total of eight reported cases and three case-series on
K. kristinae-associated infections. Most of the cases were hospital-associated infection and involved immunocompromised patients with uncontrolled diabetes, underlying cancer, underlying end-stage renal disease, prematurity,
etc. [
2345678910]. Few immunocompetent patients have been associated with this bacterial infection, but they had mostly undergone invasive procedures such as central venous catheter insertion or laparoscopic operation [
1113]. The infected sites varied, but were mostly associated with sites of catheters insertion, such as peritonitis in patients on continuous-ambulatory-peritoneal-dialysis (CAPD), bacteremia and septic pulmonary emboli in a patient with a central-venous-catheter (CVC), endocarditis in a patient with a CVC, and urinary tract infection in patient with a urinary catheter [
345811]. In some cases, genetic methods were used to confirm the pathogen identity, but in others, only automated identification systems were used for final confirmation [
3489]. Almost all cases survived, but one severe case of endocarditis in a patient with uncontrolled diabetes resulted in death [
7].
It is not uncommon to misidentify coagulase-negative staphylococcus as
Kocuria using standard biochemical analysis due to phenotypic variability. Therefore, genomic sequencing has been suggested for the precise identification of these rare microorganisms [
14]. Data are scarce regarding the reliability of
Kocuria species identification by automated systems, including the Vitek 2, Phoenix, or ATB systems [
5141516]. Recently reported cases used automation systems only for final confirmation of identification [
3489].
The limitation in our case was that we could not totally exclude the possibility of misidentification. Since a genomic identification system was not available at our hospital, and the facilities were not sufficient to preserve the organism for further examination, we were unable to perform genomic tests. Although there are some studies on other automated identification systems for the identification of
Kocuria species, no published study has reported on the MicroScan Walkaway 96 system used in our laboratory. Still, the preliminary identification provided by phenotypic tests should not be ignored [
15]. Also, it was less likely to be just a simple misidentification since the same rare organism was isolated from three different specimens.
Since the organism is part of the normal flora, there was a minor possibility of contamination, but the fact that all three sets of specimens were collected by trained health-care providers using aseptic techniques with betadine preparation and sterile equipment makes this less likely.
We hope this case may broaden the clinical spectrum of reported diseases caused by this unusual pathogen and add data on the virulence of this organism in humans.





