Abstract
Background
Targeted therapy has revolutionized the management of Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL); however, relapse still occurs because of the presence of quiescent stem cells, termed leukemia propagating cells (LPCs). This study aimed to assess the phenotypic diversity of LPCs in adult patients with Ph+ B-Acute ALL (B-ALL) and to assess its prognostic impact.
Methods
Seventy adults with newly diagnosed Ph+ B-ALL were recruited at the Mansoura Oncology Center. Multiparameter flow cytometry studies of mononuclear blast cells for cluster of differentiation (CD)34, CD38, and CD58 were performed.
Results
Seventeen patients had blasts with the pattern of LPCs (CD34+CD38−CD58−), while 53 cases had other diverse phenotypic patterns. The rate of complete response was significantly lower in patients with the LPC phenotype (47% vs. 81%, P=0.006). The median time to achieve a complete response was prolonged in patients with the CD34+CD38−CD58− phenotype (48 vs. 32 days, P=0.016). The three-year overall survival was significantly lower in patients with the CD34+CD38−CD58− phenotype (37% vs. 55% respectively, P=0.028). Multivariate analysis showed that the CD34+CD38− CD58− phenotype was an independent risk factor for overall survival.
Go to : 
Tyrosine kinase inhibitors (TKIs) are highly effective in the treatment of Philadelphia chromosome positive (Ph+) precursor B-Acute lymphoblastic leukemia (Ph+ B-ALL) [1]. TKIs improve the remission rate by >90%, increasing the overall survival rate in patients with Ph+ B-ALL to 40–50% within different risk groups [2]. However, relapse still occurs, which could be explained by the persistence of leukemia-quiescent stem cells, also known a leukemia-propagating cells (LPCs) or leukemia initiating cells. These cells may explain resistance to chemotherapy and poor long-term prognosis, in addition to disease recurrence. Novel therapeutic approaches are required to eradicate these cells [3].
LPCs are quiescent stem cells characterized by their self-renewal ability and proliferating capacity, in addition to leukemogenesis [4]. Lapidot et al. [5] reported that LPCs in acute myeloid leukemia have diverse phenotypes; however, most are clusters of differentiation (CD)34+/CD38−. Recent research reported that LPCs have a distinct gene expression profile and are associated with worse clinical outcomes, following chemotherapy [6].
The phenotypic diversity of LPCs has been studied. Kong et al. [7] that reported that CD34+CD38+CD19+, as well as CD34+CD38−CD19+, cells are self-renewing, leukemia-initiating cells. Long et al. [8] concluded that a high proportion of CD34+CD38− cells is associated with poor prognosis and inferior outcome.
CD58, also known as lymphocyte function adhesion molecules-3 (LFA-3), plays a central role in naïve and memory T helper cells during the early phase of an immune response. [9]. CD58 is reported to be overexpressed in leukemia blast cells and might be useful as minimal residual disease marker in B-ALL. Higher expression of CD58 is reported to correlate with better outcome [10]; however, in adult precursor Ph+ B-ALL, there are little data on the potential prognostic importance of differences in the CD58 expression pattern in CD34+CD38− cells. This study aimed to assess the phenotypic diversity in LPCs in adult Egyptian patients with Ph+ B-ALL and to study its prognostic impact on overall survival.
Go to : 
Seventy adults who were newly diagnosed with Ph+ B-ALL were selected from 215 patients with ALL recruited at the Mansoura Oncology Center from 2013 to 2016. Diagnosis was based on the World Health Organization criteria in a specialized hematology laboratory with International Canadian Accreditation. Routine immunophenotyping and molecular PCR for BCR, RhoGEF, and GTPase activating protein-ABL proto-oncogene 1, non-receptor tyrosine kinase (BCR-ABL) mRNA was performed. The patients were aged from 19–65 years. Inclusion criteria were Ph+ B-ALL and no contraindication for imatinib therapy; the exclusion criteria were negative for the Philadelphia chromosome, T-lineage, or childhood ALL.
The studied ALL patients were treated according to our local intuitional protocol: hyper CVAD [cyclophosphamide, vincristine, doxorubicin (trade name, Adriamycin), and dexamethasone] regimen plus imatinib [11]. However, three patients presented with poor performance status (one patient) and compromised organ functions (two patients), these patients received induction therapy with vincristine, steroids, and imatinib. Fit patients achieving a complete remission (CR) with an available matched donor had the option to proceed to allogenic stem cell transplantation (performed for 13 patients).
All procedures in the present study were performed in accordance with the ethical standards of our local institutional committee and with the 1964 Helsinki declaration. Informed consent was obtained from all the individual participants included in the study.
Immunophenotyping of mononuclear blast cells was performed using multiparameter flow cytometry with three lasers and eight colors. (BD FACS CANTO I. I FLOW Cytometry, V 33396202133, BD Biosciences, San Jose, CA, USA) using different fluorochrome probes CD58 fluorescein isothiocyanate (FITC) (BD Bioscience, 555920) CD34-phycoerythrin (PE) (lot no 6210609 BD), CD38-FITC (lot no 303504, Life Trade), with a routine panel of acute leukemia markers: CD45, terminal deoxynucleotidyltransferase (TdT), CD10, CD2, CD3, CD5, CD7, CD19, CD20, CD79a, CD117, and myeloperoxidase (MPO). The instrument was calibrated using compensated BD beads to check the laser beam and adjust the instrument settings.
Bone marrow (BM) and peripheral blood (PB) mononuclear cells were isolated using lymphocyte separation technique with Ficoll 400. Total RNA was extracted from cells using a miRNeasy Mini Kit (Qiagen, German town, MD, USA). The RNA concentration and purity was determined using a NanoDrop instrumen.
For mRNA quantification, cDNA was synthesized using a high capacity reverse transcription kit (Applied Biosystems, Foster City, CA, USA) a 20-µL reaction was prepared as follows: 2 µL of 10× RT buffer, 0.8 µL of 25× dNTPs 100 mM, 2 µL of 10× random primers, 1 µL of Multiscribe reverse transcriptase enzyme (50 U/µL), 1 µL of RNase inhibitor, 13.2 µL of nuclease free water containing the extracted RNA.
The reactions were then incubated in a thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) at 25℃ for 10 mins, 37℃ for 120 min, 85℃ for 5 min, and then held at 4℃. Real-time PCR was performed on a DNA Technology DTprime4 instrument (Prix Galien, Russia) using TaqMan gene expression assays for BCR-ABL. A 20-µL reaction was prepared as follows: 10 µL of ABI 2× universal PCR master mix, 1 µL of forward primer, and 9 µL of diluted cDNA were added. The PCR volumes were then loaded into the PCR machine and incubated at 95℃ for 10 min, followed by 40 cycles of 95℃ for 10 sec and 60℃ for 30 sec.
A conventional cytogenetic chromosomal analysis was performed by diagnosis of short-term cultures of BM specimens and standard conventional cytogenetic protocols. At least 20 metaphase chromosome samples were analyzed in each case, and the clonal abnormalities were classified according to the 2013 International System for Human Cytogenetic Nomenclature (ISCN) Guidelines [12].
Interphase fluorescent in situ hybridization (FISH) analysis was performed on BM samples using an Olympus BX61 fluorescent microscope equipped with filters suitable for fluorescein, rhodamine, and 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI). Representative images were captured via a monochrome digital camera using Cytovision Image Capture software (Applied Imaging, Grand Rapids, MI, USA). All samples were investigated using interphase FISH with the following probes: LIS BCR/ABL1 dual fusion FISH probe [t(9:22)(q34:12;q11.23)]; LIS TEL/AML1 [also known as ETS variant 6 (ETV6)/runt related transcription factor 1 (RUNX1)] dual color, dual fusion translocation probe [t(12:21) (p13;q22)]; LSI E2A [also known as transcription factor 3 (TCF3)]/PBX homeobox 1 (PBX1) dual color, dual fusion translocation [t(1;19)(q23;p13)]; and an lysine (K)-specific methyltransferase 2A (KMT2A, also known as MLL) dual color, break apart rearrangement probe the KMT2A gene at 11q23.3 rearrangement (Cytocell FISH probes, Oxford Gene Technology, Oxfords, UK). The result was considered positive if the percentage of nucleoli with abnormal signals exceeded the normal reference range (>5%) in normal cells; these probes will appear as discrete red and green spots, one for each homolog [resulting in a two green-two red (2G2R) pattern]. In cells with a translocation, there will be two yellow fusion signals in addition to the red and green signals of the normal chromosomes (1R1G, 2Y). Rearrangement-positive cases would appear with 2Y signals, while negative cases showed 2G2R or 1G1R1Y.
Data were analyzed on a personal computer running Medcalc for Windows version 15. A P-value of ≤0.05 was considered statistically significant. Association between categorical variables was tested using the chi-square test. Fisher's exact test was used if the assumptions of the chi-square test were violated. The independent sample t test was used to compare the means between two groups. For nonparametric analysis, the Mann–Whitney U test was used. Survival was calculated using the Kaplan–Meier Product-Limit Estimator. Survival comparisons were performed using the log-rank test to explore variables for their independent prognostic effect on survival; this was performed using the multivariate Cox's proportional regression hazard model.
Go to : 
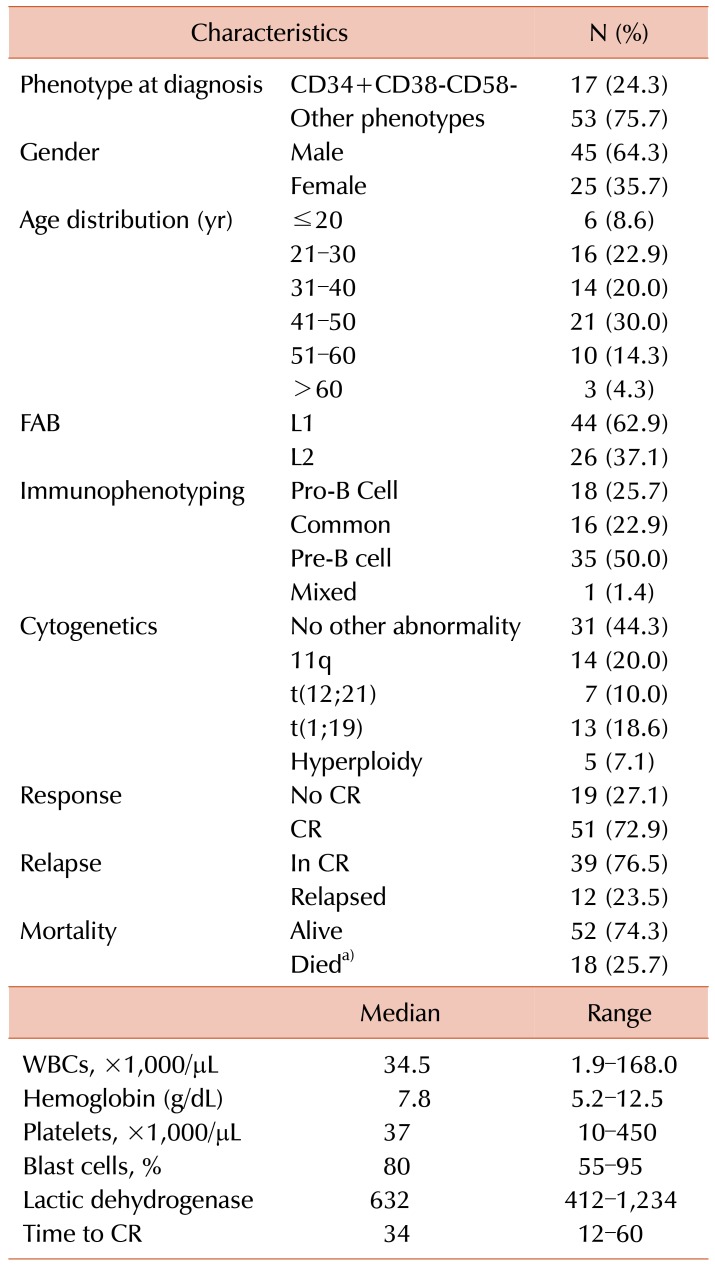
In our cohort study, 70 patients with precursor positive Philadelphia chromosome B-ALL were selected from the Oncology Center at Mansoura University. Their ages ranged from 19–65 years, with a median of 41 years. Morphological assessment revealed that 62.9% of the cases were French American British classification (FAB) L1, and half of them were suffering from pre-B-ALL. Their cytogenetic features were: 20% associated with MLL rearrangement 11q, 18.6% associated with t(1:19), 10% associated with t(12:21), 7.1% were hyperploid, and no other specific abnormalities were present in 44.3%.
The majority of the patients in the present study (72.9%) achieved CR; however, follow up for 36 months showed that 23.5% relapsed.
The immunophenotyping pattern for the gating strategy in the CD34+ blast population depended on a cut off value of 20% or more for CD38+, while a lower percentage was regarded as negative. Furthermore, in the CD34+CD38− population, a CD58+ cut off of 20% or more was used; a score of less than 20% was considered as negative, as described by Kong et al. [13].
Seventeen patients had a pattern consistent with LPCs: CD34+CD38−CD58−, while 53 cases have diverse phenotypic patterns, such as CD34+CD38+CD58+ and CD38−CD58+, CD38+CD58−, as presented in Table 1.
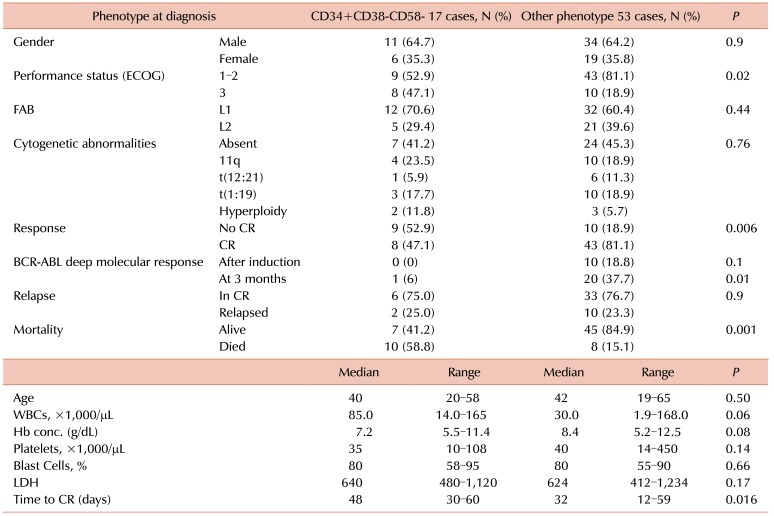
Comparison of LPCs with other phenotypic groups revealed no significant difference in age and sex, leukocyte count, hemoglobin (Hb) concentration, platelet count, blast percentage in the BM, and lactate dehydrogenase (LDH) levels. Patients with the LPC phenotype had significantly worse performance status score according to the Eastern Cooperative Oncology Group (ECOG) classification: 52.9% of the patients with CD34+CD38−CD58− were of performance status 1–2 and 47.1% scored 3. In the other phenotype groups, 81.1% showed a good performance status of 1–2, with a moderately significant difference between the two groups (LPC phenotype vs. other phenotypes, P=0.02) as illustrated in Table 2.
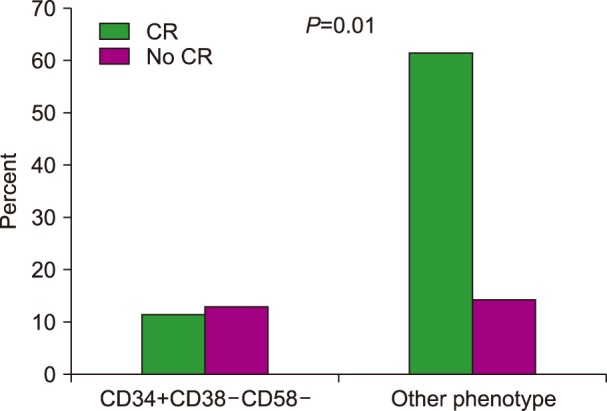
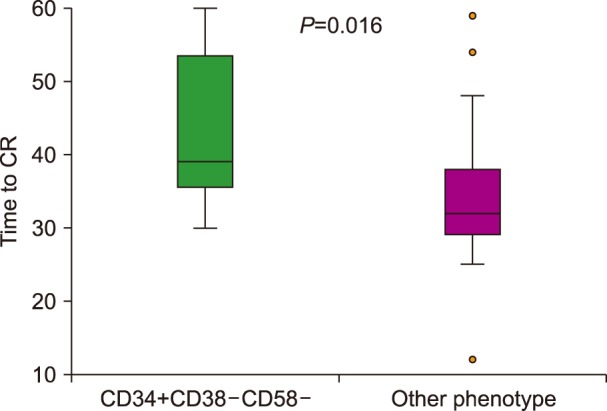
The rate of complete response was significantly lower in patients with the LPC phenotype (47% vs. 81%, P=0.006), as presented in Fig. 1. The median time to achieve a complete response was prolonged in patients with the CD34+CD38−CD58− phenotype (48 vs. 32 days, P=0.016), as shown in Fig. 2. In addition, the deep molecular response of BCR-ABL was significantly lower in patients with the CD34+CD38−CD58− phenotype at remission after consolidation chemotherapy (P=0.01).
Response to chemotherapy was not good in patients with the LPC phenotype: 75% underwent further incomplete remission and 25% underwent relapse, with no significant difference compared with that in patients with other phenotypes. Mortality was higher in patients with the LPC phenotype compared with that in patients with other phenotypes (58.8% vs. 15.1%, P=0.001).
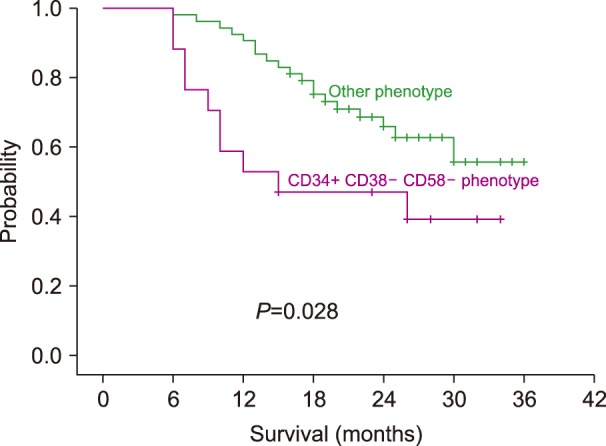
Survival analysis using the Kaplan–Meir method revealed a shorter survival time for patients with the CD34+CD38−CD58− phenotype than for patients with other phenotypes. The median survival time for patients with the CD34+CD38−CD58− phenotype was 15 months [95% confidence interval (CI), 11–25 mo]; the median survival for patients with other phenotypes was not reached. The three-year overall survival was significantly lower in patients with the CD34+CD38−CD58− phenotype (37% vs. 55% respectively, log rank 4.8, P=0.028).
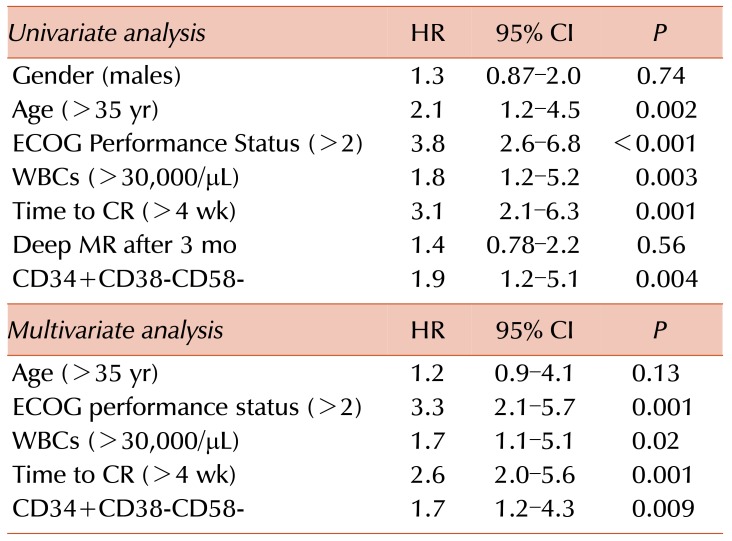
Multivariate analysis showed that the CD34+CD38−CD58− phenotype was an independent risk factor (Hazard Ratio (HR)=1.7, 95% CI=1.2–4.3; P=0.009), as well as performance status (HR=3.3, 95% CI=2.1–5.7), and time to complete response (HR=2.6, 95% CI=2.0–5.6). Among the studied parameters, age >35 years, poor performance status, high white blood cells (WBCs) count (>30,000/µL), longer time to complete remission (>4 wk), and the CD34+CD38−CD58− phenotype were prognostic factors for overall survival at the univariate analysis. In multivariate analysis, poor performance status, a high WBC count (>30,000/µL), longer time to CR (>4 wk), and the CD34+CD38− CD58− phenotype were independent prognostic factors of overall survival, as shown in Fig. 3 and Table 3.
Go to : 
Precursor B-Acute lymphoblastic leukemia (Pre-B ALL) with the characteristic Philadelphia chromosome-positive translocation in adults is a highly aggressive hematological malignancy of the BM originating from immature lymphoid cells at the early stages of differentiation. Pre-B ALL is associated with poor survival, and high morbidity and mortality [14]. Recently, targeted therapy using TKIs, in addition to other chemotherapy, has improved the outcome by up to 90%; however, relapse still occurs. This could be explained by the presence of specific quiescent stem cells with a characteristic phenotype, the LPCs [4]. However, the existence, phenotype, biological properties, and clinical significance of these LPCs remain highly controversial. Therefore, we aimed to assess these cells using multiparameter flow cytometry, in association with their clinicopathological features and prognostic impact.
The main finding of our study was that 24.3% of the patients with Ph+ ALL had a specific CD34+CD38−CD58− phenotype. We did not detect any variation in this percentage; only one similar study by Kong et al. [13] showed that 20.6% of the patients had this phenotype.
The present study is the first to assess the relationship between the CD34+CD38−CD58− phenotype and the ALL morphological pattern. This phenotype was associated with French American British classification (FAB) L1 in 70.6% of the cases. FAB L1 is associated with bad prognosis, especially when diagnosed as undifferentiated mixed clonality leukemia.
Demographic and laboratory data did not show significant differences between the two phenotypic groups regarding age, gender, Hb, WBCs, blast percentage in the BM, platelet count, and LDH. However, a poor clinical performance status score of 2 or more was associated with the CD34+CD38−CD58− phenotype. This finding did not agree with that of Long et al. [8], who revealed that a percentage of CD34+CD38− cells was associated with clinical and biological features of ALL patients. This could be explained by the fact that the phenotypic characteristics of cells differed between the two studies. In addition, analyzing the CD58 status in our population might have generated a statistical difference between the two phenotypes in our study.
Response to induction therapy was slower in patients with the LPC phenotype than in patients with the other phenotypes. Median time to achieve complete response was significantly prolonged compared with that in patients with other phenotypic patterns. This finding agreed with that of Kong et al. [13].
Association of CD34+CD38−CD58− with the MLL rearrangement was associated with poor prognosis in some cases; however, this finding did not reach statistical significance. This observation corresponded with the findings of Aoki et al. [15], who identified CD34+ leukemia-initiating cells in MLL rearranged ALL.
Incomplete remission with relapse increased in patients with the CD34+CD38−CD58− phenotype but did not reach statistical significant. This agreed with the findings of Long et al. [8], who found that a high proportion of patients with a CD34+CD38− phenotype showed poor prognosis of childhood ALL and increased relapse with a negative correlation to their outcome. However, Kong et al. [13] revealed that the cumulative incidence of relapse was statistically higher for the CD34+CD38−CD58− phenotype than for other phenotypes.
The three-year overall survival was significantly shorter in the CD34+CD38−CD58− group than in the other groups. This finding agreed with that of Kong et al. [13], who reported that the CD34+CD38−CD58−cohort had a worse 3-year survival rate compared with that in the other phenotype cohorts; however, the difference was not significant in that study. Archimbaud et al. [10] found a correlation between lower CD58 expression on ALL blasts and worse survival. We found inferior outcome and poor prognosis in the CD34+CD38−CD58− group compared with that in the other phenotype groups. In addition Kong et al. [16] found that in a cohort of patients with allogenic bone marrow transplantation, the presence of CD34+CD38−CD58− LPCs correlated directly with higher 3-year cumulative incidence of relapse, and worse leukemia free-survival and OS.
Multivariate analysis revealed that an ECOG performance status score of 2 or more, a higher leukocytic count and time to complete response (more than 4 wk), as well as a CD34+CD38−CD58− phenotype, could be independent risk factors for a worse outcome. This agreed with the results of Kong et al. [13].
A recent study reported that potential cancer stem cells have specific cell expression and a leukemogenesis effect [17]. Thalheimer et al. [18] highlighted the heterogeneity of individual ALL cells with variable abilities to engraft and progress to leukemia in NOD scid gamma mice and proposed the existence of LPCs in B-cell percursor-ALL. These cells are pivotal and are a prerequisite for the development of LPCs-directed therapies. Novel strategies are urgently required to improve the long term outcome and improve survival in patients with Ph+ B-ALL [19].
Finally, our data suggest that the presence of CD34+CD38−CD58− LPCs at diagnosis would allow the rapid identification of higher risk patients. Risk stratification of these patients could be used to further guide therapy and develop effective LPCs-targeted strategies to improve treatment outcome.
Go to : 
Notes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
Go to : 
References
1. Ottmann OG, Pfeifer H. Management of Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL). Hematology Am Soc Hematol Educ Program. 2009; 371–381. PMID: 20008223.

2. Bassan R, Hoelzer D. Modern therapy of acute lymphoblastic leukemia. J Clin Oncol. 2011; 29:532–543. PMID: 21220592.

3. Gökbuget N, Hoelzer D. Treatment of adult acute lymphoblastic leukemia. Semin Hematol. 2009; 46:64–75. PMID: 19100369.

4. Cobaleda C, Gutiérrez-Cianca N, Pérez-Losada J, et al. A primitive hematopoietic cell is the target for the leukemic transformation in human Philadelphia-positive acute lymphoblastic leukemia. Blood. 2000; 95:1007–1013. PMID: 10648416.

5. Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994; 367:645–648. PMID: 7509044.

6. Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011; 17:1086–1093. PMID: 21873988.

7. Kong Y, Yoshida S, Saito Y, et al. CD34+CD38+CD19+ as well as CD34+CD38−CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia. 2008; 22:1207–1213. PMID: 18418410.

8. Long J, Liu S, Li K, Zhou X, Zhang P, Zou L. High proportion of CD34+/CD38−cells is positively correlated with poor prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2014; 55:611–617. PMID: 23706103.

9. Wang JH, Smolyar A, Tan K, et al. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999; 97:791–803. PMID: 10380930.

10. Archimbaud E, Thomas X, Campos L, Magaud JP, Doré JF, Fiere D. Expression of surface adhesion molecules CD54 (ICAM-1) and CD58 (LFA-3) in adult acute leukemia: relationship with initial characteristics and prognosis. Leukemia. 1992; 6:265–271. PMID: 1375302.
11. Thomas DA, Faderl S, Cortes J, et al. Treatment of Philadelphia chromosome-positive acute lymphocytic leukemia with hyper-CVAD and imatinib mesylate. Blood. 2004; 103:4396–4407. PMID: 14551133.

12. Shaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Basel, Switzerland: S. Karger;2013.
13. Kong Y, Chang YJ, Liu YR, et al. CD34(+)CD38(−)CD58(−) cells are leukemia-propagating cells in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia. 2014; 28:2398–2401. PMID: 25135692.

14. le Viseur C, Hotfilder M, Bomken S, et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell. 2008; 14:47–58. PMID: 18598943.

15. Aoki Y, Watanabe T, Saito Y, et al. Identification of CD34+ and CD34− leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia. Blood. 2015; 125:967–980. PMID: 25538041.

16. Kong Y, Xu LP, Liu YR, et al. Presence of CD34(+)CD38(−)CD58(−) leukemia-propagating cells at diagnosis identifies patients at high risk of relapse with Ph chromosome-positive ALL after all-ohematopoietic SCT. Bone Marrow Transplant. 2015; 50:348–353. PMID: 25486581.

17. Gunjal P, Pedziwiatr D, Ismail AA, Kakar SS, Ratajczak MZ. An emerging question about putative cancer stem cells in established cell lines-are they true stem cells or a fluctuating cell phenotype? J Cancer Stem Cell Res. 2015; 3:pii:e1004.

18. Thalheimer FB, Wingert S, De Giacomo P, et al. Cytokine-regulated GADD45G induces differentiation and lineage selection in hematopoietic stem cells. Stem Cell Reports. 2014; 3:34–43. PMID: 25068120.

19. Lang F, Wojcik B, Bothur S, et al. Plastic CD34 and CD38 expression in adult B-cell precursor acute lymphoblastic leukemia explains ambiguity of leukemia-initiating stem cell populations. Leukemia. 2017; 31:731–734. PMID: 27956738.

Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download