INTRODUCTION
DOES THE GUT MICROBIOME HAVE AN IMPACT ON AD?
Gut microbiome and AD
Table 1
Summary of the previous studies of the gut microbiome in AD
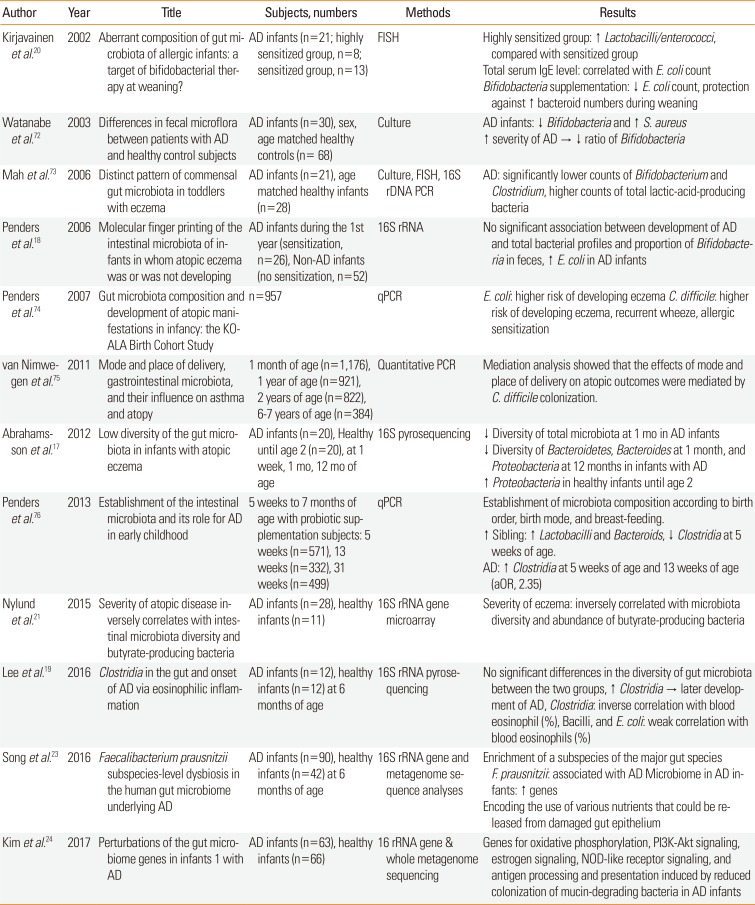
| Author | Year | Title | Subjects, numbers | Methods | Results |
|---|---|---|---|---|---|
| Kirjavainen et al.20 | 2002 | Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? | AD infants (n=21; highly sensitized group, n=8; sensitized group, n=13) | FISH | Highly sensitized group: ↑ Lactobacilli/enterococci, compared with sensitized group |
| Total serum IgE level: correlated with E. coli count | |||||
| Bifidobacteria supplementation: ↓ E. coli count, protection against ↑ bacteroid numbers during weaning | |||||
| Watanabe et al.72 | 2003 | Differences in fecal microflora between patients with AD and healthy control subjects | AD infants (n=30), sex, age matched healthy controls (n= 68) | Culture | AD infants: ↓ Bifidobacteria and ↑ S. aureus ↑ severity of AD → ↓ ratio of Bifidobacteria |
| Mah et al.73 | 2006 | Distinct pattern of commensal gut microbiota in toddlers with eczema | AD infants (n=21), age matched healthy infants (n=28) | Culture, FISH, 16S rDNA PCR | AD: significantly lower counts of Bifidobacterium and Clostridium, higher counts of total lactic-acid-producing bacteria |
| Penders et al.18 | 2006 | Molecular finger printing of the intestinal microbiota of infants in whom atopic eczema was or was not developing | AD infants during the 1st year (sensitization, n=26), Non-AD infants (no sensitization, n=52) | 16S rRNA | No significant association between development of AD and total bacterial profiles and proportion of Bifidobacteria in feces, ↑ E. coli in AD infants |
| Penders et al.74 | 2007 | Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study | n=957 | qPCR | E. coli: higher risk of developing eczema C. difficile: higher risk of developing eczema, recurrent wheeze, allergic sensitization |
| van Nimwe-gen et al.75 | 2011 | Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy | 1 month of age (n=1,176), 1 year of age (n=921), 2 years of age (n=822), 6-7 years of age (n=384) | Quantitative PCR | Mediation analysis showed that the effects of mode and place of delivery on atopic outcomes were mediated by C. difficile colonization. |
| Abrahamsson et al.17 | 2012 | Low diversity of the gut microbiota in infants with atopic eczema | AD infants (n=20), Healthy until age 2 (n=20), at 1 week, 1 mo, 12 mo of age | 16S pyrosequencing | ↓ Diversity of total microbiota at 1 mo in AD infants |
| ↓ Diversity of Bacteroidetes, Bacteroides at 1 month, and Proteobacteria at 12 months in infants with AD | |||||
| ↑ Proteobacteria in healthy infants until age 2 | |||||
| Penders et al.76 | 2013 | Establishment of the intestinal microbiota and its role for AD in early childhood | 5 weeks to 7 months of age with probiotic supplementation subjects: 5 weeks (n=571), 13 weeks (n=332), 31 weeks (n=499) | qPCR | Establishment of microbiota composition according to birth order, birth mode, and breast-feeding. |
| ↑ Sibling: ↑ Lactobacilli and Bacteroids, ↓ Clostridia at 5 weeks of age. | |||||
| AD: ↑ Clostridia at 5 weeks of age and 13 weeks of age (aOR, 2.35) | |||||
| Nylund et al.21 | 2015 | Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria | AD infants (n=28), healthy infants (n=11) | 16S rRNA gene microarray | Severity of eczema: inversely correlated with microbiota diversity and abundance of butyrate-producing bacteria |
| Lee et al.19 | 2016 | Clostridia in the gut and onset of AD via eosinophilic inflammation | AD infants (n=12), healthy infants (n=12) at 6 months of age | 16S rRNA pyrosequencing | No significant differences in the diversity of gut microbiota between the two groups, ↑ Clostridia → later development of AD, Clostridia: inverse correlation with blood eosinophil (%), Bacilli, and E. coli: weak correlation with blood eosinophils (%) |
| Song et al.23 | 2016 | Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying AD | AD infants (n=90), healthy infants (n=42) at 6 months of age | 16S rRNA gene and metagenome sequence analyses | Enrichment of a subspecies of the major gut species F. prausnitzii: associated with AD Microbiome in AD infants: ↑ genes |
| Encoding the use of various nutrients that could be released from damaged gut epithelium | |||||
| Kim et al.24 | 2017 | Perturbations of the gut microbiome genes in infants 1 with AD | AD infants (n=63), healthy infants (n=66) | 16 rRNA gene & whole metagenome sequencing | Genes for oxidative phosphorylation, PI3K-Akt signaling, estrogen signaling, NOD-like receptor signaling, and antigen processing and presentation induced by reduced colonization of mucin-degrading bacteria in AD infants |
AD, atopic dermatitis; FISH, fluorescence in situ hybridization; E. coli, Escherichia coli; IgE, immunoglobulin E; S. aureus, Staphylococcus aureus; rDNA, recombinant DNA; rRNA, ribosomal ribonucleic acid; qPCR, quantitative polymerase chain reaction; aOR, adjusted odds ratio; NOD, nucleotide-binding oligomerization domain.
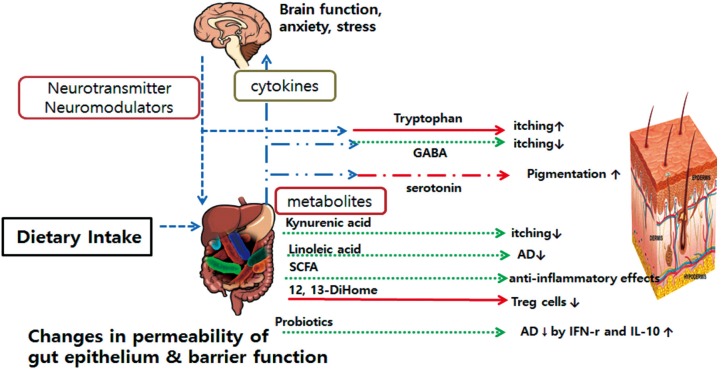
HOW DO GUT MICROBIOME IMPACT AD?
Immunologic pathway
Metabolite pathway
Neuroendocrine pathway
HOW DOES SKIN IMMUNITY IMPACT ON SYSTEMIC IMMUNITY AND AD?
Skin microbiome and AD
Table 2
Summary of the previous studies on the skin microbiome in AD
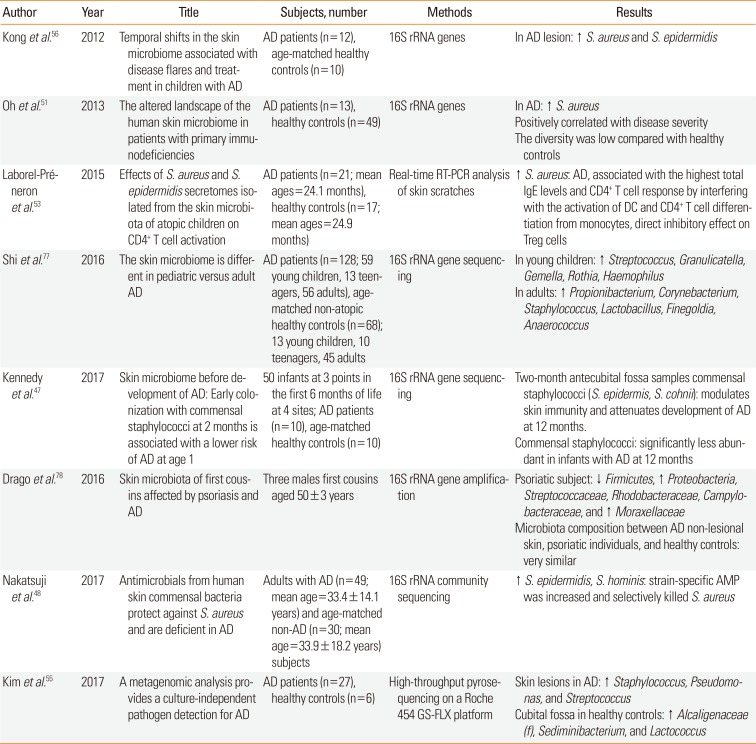
| Author | Year | Title | Subjects, number | Methods | Results |
|---|---|---|---|---|---|
| Kong et al.56 | 2012 | Temporal shifts in the skin microbiome associated with disease flares and treatment in children with AD | AD patients (n=12), age-matched healthy controls (n=10) | 16S rRNA genes | In AD lesion: ↑ S. aureus and S. epidermidis |
| Oh et al.51 | 2013 | The altered landscape of the human skin microbiome in patients with primary immunodeficiencies | AD patients (n=13), healthy controls (n=49) | 16S rRNA genes | In AD: ↑ S. aureus |
| Positively correlated with disease severity | |||||
| The diversity was low compared with healthy controls | |||||
| Laborel-Préneron et al.53 | 2015 | Effects of S. aureus and S. epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation | AD patients (n=21; mean ages=24.1 months), healthy controls (n=17; mean ages=24.9 months) | Real-time RT-PCR analysis of skin scratches | ↑ S. aureus: AD, associated with the highest total IgE levels and CD4+ T cell response by interfering with the activation of DC and CD4+ T cell differentiation from monocytes, direct inhibitory effect on Treg cells |
| Shi et al.77 | 2016 | The skin microbiome is different in pediatric versus adult AD | AD patients (n=128; 59 young children, 13 teenagers, 56 adults), age-matched non-atopic healthy controls (n=68); 13 young children, 10 teenagers, 45 adults | 16S rRNA gene sequencing | In young children: ↑ Streptococcus, Granulicatella, Gemella, Rothia, Haemophilus |
| In adults: ↑ Propionibacterium, Corynebacterium, Staphylococcus, Lactobacillus, Finegoldia, Anaerococcus | |||||
| Kennedy et al.47 | 2017 | Skin microbiome before development of AD: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of AD at age 1 | 50 infants at 3 points in the first 6 months of life at 4 sites; AD patients (n=10), age-matched healthy controls (n=10) | 16S rRNA gene sequencing | Two-month antecubital fossa samples commensal staphylococci (S. epidermis, S. cohnii): modulates skin immunity and attenuates development of AD at 12 months. |
| Commensal staphylococci: significantly less abundant in infants with AD at 12 months | |||||
| Drago et al.78 | 2016 | Skin microbiota of first cousins affected by psoriasis and AD | Three males first cousins aged 50±3 years | 16S rRNA gene amplification | Psoriatic subject: ↓ Firmicutes, ↑ Proteobacteria, Streptococcaceae, Rhodobacteraceae, Campylobacteraceae, and ↑ Moraxellaceae |
| Microbiota composition between AD non-lesional skin, psoriatic individuals, and healthy controls: very similar | |||||
| Nakatsuji et al.48 | 2017 | Antimicrobials from human skin commensal bacteria protect against S. aureus and are deficient in AD | Adults with AD (n=49; mean age=33.4±14.1 years) and age-matched non-AD (n=30; mean age=33.9±18.2 years) subjects | 16S rRNA community sequencing | ↑ S. epidermidis, S. hominis: strain-specific AMP was increased and selectively killed S. aureus |
| Kim et al.55 | 2017 | A metagenomic analysis provides a culture-independent pathogen detection for AD | AD patients (n=27), healthy controls (n=6) | High-throughput pyrosequencing on a Roche 454 GS-FLX platform | Skin lesions in AD: ↑ Staphylococcus, Pseudomonas, and Streptococcus |
| Cubital fossa in healthy controls: ↑ Alcaligenaceae (f), Sediminibacterium, and Lactococcus |
AD, atopic dermatitis; rRNA, ribosomal ribonucleic acid; S. aureus, Staphylococcus aureus; S. epidermidis, Staphylococcus epidermidis; RT-PCR, reverse transcription polymerase chain reaction; IgE, immunoglobulin E; DC, dendritic cell; Treg, regulatory T; S. cohnii, Staphylococcus cohnii; S. hominis, Staphylococcus hominis.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download