The 21-gene expression Oncotype DX™ (ODX) assay (Genomic Health, Redwood City, USA) is available for patients with estrogen receptor (ER)-positive/human epidermal growth factor receptor 2 (HER2)-negative/lymph node-negative disease. Three risk groups were defined (low, intermediate, and high) with recurrence score (RS) of <18, 18–30, and >30, respectively, corresponding to a 10-year distant recurrence risk of <10%, 10%–20%, and >20%, respectively [
1]. The ODX test predicted that patients in the high-risk group would experience greater benefits from chemotherapy and patients in the low-risk group would experience no benefit from chemotherapy over endocrine therapy alone; however, the benefit of chemotherapy for patients in the intermediate group was uncertain [
23]. The Trial Assigning Individualized Options for Treatment (TAILORx) was established in 2006. In the TAILORx trial, the low-risk group included patients with RS of <11, the intermediate-risk group included those with RS of 11–25, and the high-risk group included those with RS of >25 [
4].
ODX testing is currently endorsed by the American Society of Clinical Oncology and the National Comprehensive Cancer Network [
56]. However, ODX testing is expensive (the current estimated cost is ~$4,000), implying that it is performed in only about one-third of patients with breast cancer who meet the inclusion criteria in the United States [
7] and in <20% of eligible patients in European countries [
8].
A recent study conducted at the University of Tennessee Medical Center aimed to predict high- or low-risk ODX RS test results based on clinicopathologic variables. The researchers developed a nomogram based on a large dataset of ODX-tested female patients with ER-positive/HER2-negative/lymph node-negative disease and invasive breast carcinomas sized between 6 and 50 mm from the National Cancer Database (NCDB). The nomogram used six clinicopathologic variables: age, tumor size, tumor grade, progesterone receptor status, lymphovascular invasion, and histologic breast cancer type (among the four most common types). In their study, 27,685 original cohorts and 12,763 external validation cohorts were used to develop nomograms, and the nomograms showed high, acceptable concordance indices (c-indices) measured by the receiver operating characteristic (ROC) curve. The c-indices for commercial and TAILORx ODX cutoff values was 0.89 (95% confidence interval [CI], 0.88–0.90) and 0.852 (95% CI, 0.842–0.861), respectively. An online nomogram calculator, which could calculate the probability of low- or high-risk ODX RSs for each patient, was provided by the University of Tennessee Medical Center on its website [
9].
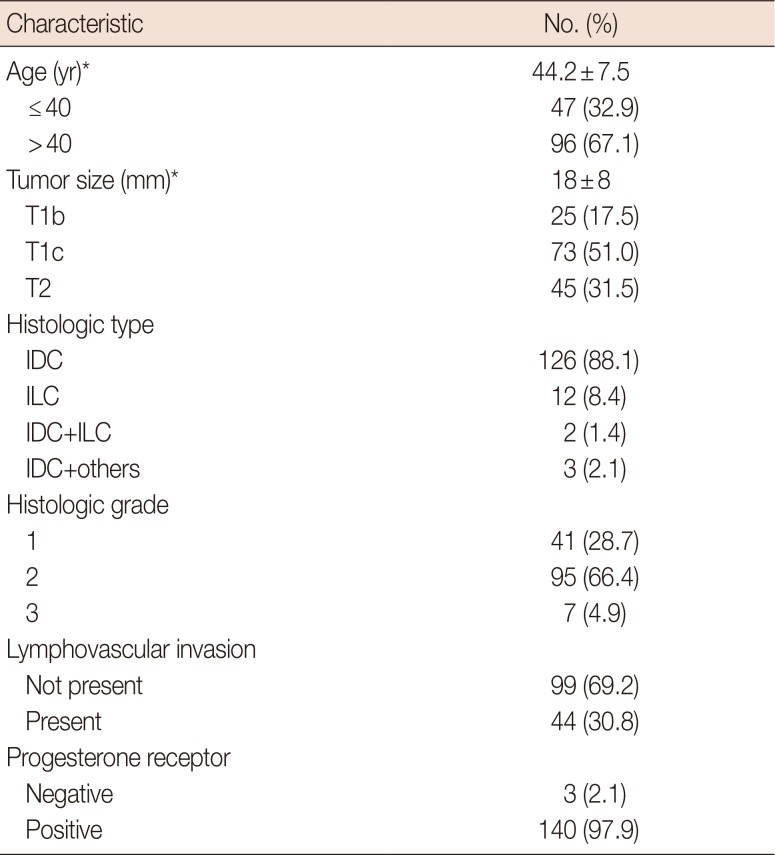
We performed a retrospective review of 218 patients who underwent breast-conserving surgery or mastectomy at a single institution and ODX testing between April 2011 and January 2017 to confirm that the nomogram results were consistent with our data, which differed from those of the NCDB in terms of race and ethnicity. Among the 218 patients, 68 had lymph node metastasis, three had HER2-positive breast cancer, five had tumors smaller than 5 mm, and one had a tumor greater than 50 mm in size. Seven patients had no information about the six clinicopathologic variables used in the nomogram. Some patients exhibited multiple exclusion criteria, so a total of 143 patients were eligible for the nomogram. The clinicopathological characteristics of the 143 patients are shown in
Table 1. Most patients were in the low-risk group. The distribution was 95 (66.4%) in the low-risk group, 43 (30.1%) in the intermediate-risk group, and 5 (3.5%) in the high-risk group for commercial cutoff values; and 28 (19.6%) in the low-risk group, 101 (70.6%) in the intermediate-risk group, and 14 (9.8%) in the high-risk group for TAILORx cutoff values.
Table 1
Clinicopathologic characteristics of 143 patients
|
Characteristic |
No. (%) |
|
Age (yr)*
|
44.2±7.5 |
|
≤40 |
47 (32.9) |
|
>40 |
96 (67.1) |
|
Tumor size (mm)*
|
18±8 |
|
T1b |
25 (17.5) |
|
T1c |
73 (51.0) |
|
T2 |
45 (31.5) |
|
Histologic type |
|
|
IDC |
126 (88.1) |
|
ILC |
12 (8.4) |
|
IDC+ILC |
2 (1.4) |
|
IDC+others |
3 (2.1) |
|
Histologic grade |
|
|
1 |
41 (28.7) |
|
2 |
95 (66.4) |
|
3 |
7 (4.9) |
|
Lymphovascular invasion |
|
|
Not present |
99 (69.2) |
|
Present |
44 (30.8) |
|
Progesterone receptor |
|
|
Negative |
3 (2.1) |
|
Positive |
140 (97.9) |

The nomograms were validated by discrimination and calibration. Discrimination was assessed using ROC curves to yield c-indices, which estimate the probability of concordance between the predicted and observed results. The concordance index (c-index) can range from 0.5 to 1.0, indicating random predictions and perfect concordance, respectively. We also calculated the sensitivity, specificity, and positive predictive value (PPV) based on cutoff values determined by the Youden index. The calibration plot was applied to assess the prediction accuracy of the nomograms by plotting the actual result against the nomogram-predicted probabilities. All statistical analyses were conducted using SAS version 9.4 (SAS Institute Inc., Cary, USA).
The calibration plot of the nomograms is shown in
Figure 1. The calibration plot showed that the University of Tennessee Medical Center nomogram overestimated the probabilities of belonging in high- and low-risk groups according to commercial or TAILORx cutoff values. The predictive accuracy of the nomogram at each cutoff value is shown in
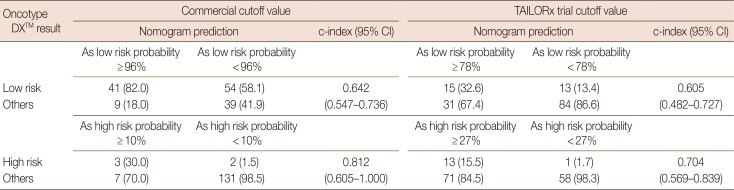
Table 2. When we applied the 96% cutoff value for the nomogram to predict membership in the low-risk commercial ODX RS group, the c-index was 0.642 (95% CI, 0.547–0.736) and its PPV was 82.0%. With the 78% cutoff predictive value by nomogram for the low-risk TAILORx RS group, the c-index was 0.605 (95% CI, 0.482–0.727) and the nomogram's PPV was 32.6%. Inspection of the high-risk commercial ODX RS group with the 10% cutoff predictive value yielded a c-index of 0.812 (95% CI, 0.605–1.000) and PPV of 30.0%. For the high-risk TAILORx RS group, the c-index was 0.704 (95% CI, 0.569–0.839) and PPV was 15.5% with the cutoff value as 27% of nomogram probability. The sensitivity of the nomogram for the low-risk commercial ODX RS group was 43.2%; the specificity of that group was 81.3%. The sensitivity for the low-risk group based on the TAILORx RS was 53.6%; the specificity was 73.0%. For the high-risk group, the sensitivity was 60.0% and specificity was 94.9% for the commercial ODX RS, and the sensitivity was 92.9% and specificity was 45.0% for the TAILORx RS.
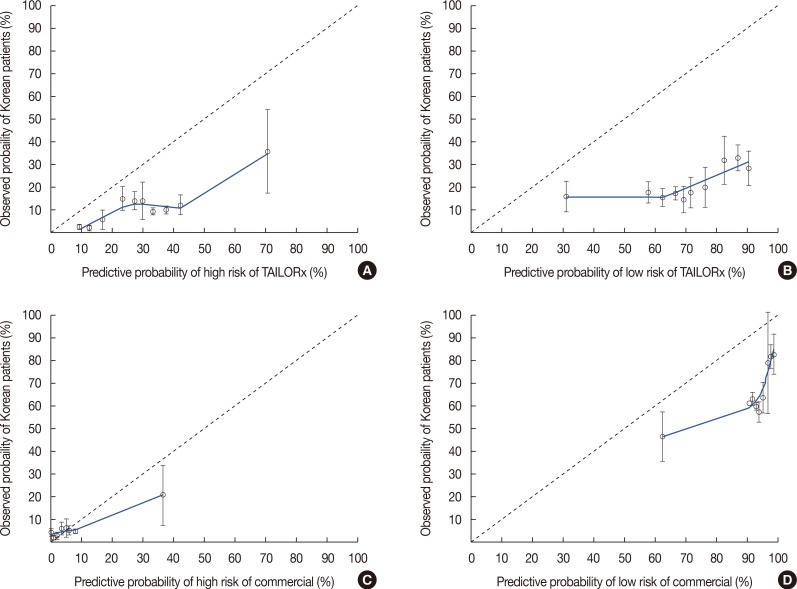 | Figure 1The calibration plots of four nomograms predicting probabilities of belonging to high- and low-risk group according to commercial and Trial Assigning Individualized Options for Treatment (TAILORx) trial criteria. (A) Nomogram to predict for high risk group according to TAILORx criteria. (B) Nomogram to predict for low risk group according to TAILORx criteria. (C) Nomogram to predict for high risk group according to commercial criteria. (D) Nomogram to predict for low risk group according to commercial criteria.
|
Table 2
Accuracy of high- and low-risk group prediction with nomograms according to the cutoff value determined by the receiver operating characteristic curve
|
Oncotype DX™ result |
Commercial cutoff value |
TAILORx trial cutoff value |
|
Nomogram prediction |
c-index (95% CI) |
Nomogram prediction |
c-index (95% CI) |
Low risk
Others |
As low risk probability ≥ 96% |
As low risk probability < 96% |
|
As low risk probability ≥ 78% |
As low risk probability < 78% |
|
|
41 (82.0) |
54 (58.1) |
0.642 |
15 (32.6) |
13 (13.4) |
0.605 |
|
9 (18.0) |
39 (41.9) |
(0.547–0.736) |
31 (67.4) |
84 (86.6) |
(0.482–0.727) |
|
As high risk probability ≥ 10% |
As high risk probability < 10% |
|
As high risk probability ≥ 27% |
As high risk probability < 27% |
|
High risk
Others |
3 (30.0) |
2 (1.5) |
0.812 |
13 (15.5) |
1 (1.7) |
0.704 |
|
7 (70.0) |
131 (98.5) |
(0.605–1.000) |
71 (84.5) |
58 (98.3) |
(0.569–0.839) |

Even though the ODX RS assay can predict the benefits of adjuvant chemotherapy in patients with early-stage breast cancer, the ODX test has been even less popular in East Asia, including Korea [
10]. The gross domestic product (GDP) per capita of Korea was $26,929 in 2015 [
11], whereas the GDP per capita of the United States was $55,836 [
12]. Thus, the ODX test is even more expensive in Korea, which could explain the less frequent ODX testing in Korea.
We applied the data from our institution to determine whether the nomograms based on the large dataset are suitable for use in Asian patients. Although the predictive accuracy of the nomogram was approximately 90% for commercial ODX RS groups and 85% for TAILORx RS groups in the study of the University of Tennessee Medical Center [
9], the c-index for a commercial ODX RS in the low-risk group in our study was much lower than that of the University of Tennessee Medical Center (0.642 vs. 0.890), and the c-index for the highrisk group was 0.812. For the TAILORx RS with our data, the c-index for the low-risk group was 0.605, and that for the high-risk group was 0.704.
Several factors could explain these discrepancies. First, the mean age of patients in this study (44.2±7.5 years), which differed from that in the University of Tennessee Medical Center study (59.3±10.3 years) [
9]. In our data, only four patients were older than 60 years. Furthermore, only 3% of the patients in the original study were younger than 40 years, whereas 32.9% (47 of 143) of the patients in our study were younger than 40 years. Partridge et al. [
13] reported that young age was a significant negative prognostic factor in women with luminal breast cancers. The significant difference in mean age between these two studies might affect the accuracy of their respective results.
Ethnic differences might also contribute to nomogram accuracy. Although several studies have shown that ODX RSs do not vary significantly by race/ethnicity [
1415], other studies showed ethnic differences in tumor biology and biomarker patterns. Albain et al. [
16] and Guth et al. [
17] reported a significantly higher expression of proliferation genes by specific ethnicity, and the correlation between high ODX RSs and high Ki-67 indices has been well established.
In the high-risk group, the c-index was relatively higher than that in the low-risk group in our data, although it was still lower than that in the University of Tennessee Medical Center's data. In addition, only five patients (3.5%) of our data were included in the high-risk group according to commercial criteria, and 14 patients (9.8%) were included in the high-risk group according to TAILORx criteria. The cutoff predictive value for validation of the nomogram determined by the Youden index was unacceptably low, which might be due to an insufficient number of patients in the high-risk RS group. Therefore, nomograms for high-risk group prediction could not be validated appropriately with our data.
In the 8th edition of the American Joint Commission on Cancer, patients with hormone receptor-positive/HER2-negative/lymph node-negative breast cancer with low-risk RSs derived from multigene breast cancer prognostic panels, such as the ODX test, have been placed into the same prognostic category as that of patients with stage I cancer, regardless of tumor size [
9]. Thus, the usefulness of the ODX test has been emphasized. Unfortunately, the ODX test remains expensive and unavailable to many patients with breast cancer in most countries. Therefore, a precise nomogram that could predict groups at ODX risk is still necessary. Although the novel nomogram for predicting ODX scores developed by researchers in the University of Tennessee Medical Center was based on a large dataset, it could not be generalized to patients in Asia. Thus, further studies using large datasets from different ethnicities should be conducted to develop a nomogram applicable to patients worldwide.







 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download