INTRODUCTION
METHODS
Ethics statement
Clinical samples
Immunohistochemical staining
Immunohistochemical evaluation
Cell culture
Plasmids, small interfering RNAs, and the overexpression vector
Real-time reverse transcription polymerase chain reaction
Western blotting analysis
MTT assay
Glucose-uptake, lactate, adenosine triphosphate, and lactate dehydrogenase assays
Immunofluorescence assay
Chromatin immunoprecipitation
Statistical analysis
RESULTS
The level of HDAC3 expression was upregulated in breast cancer
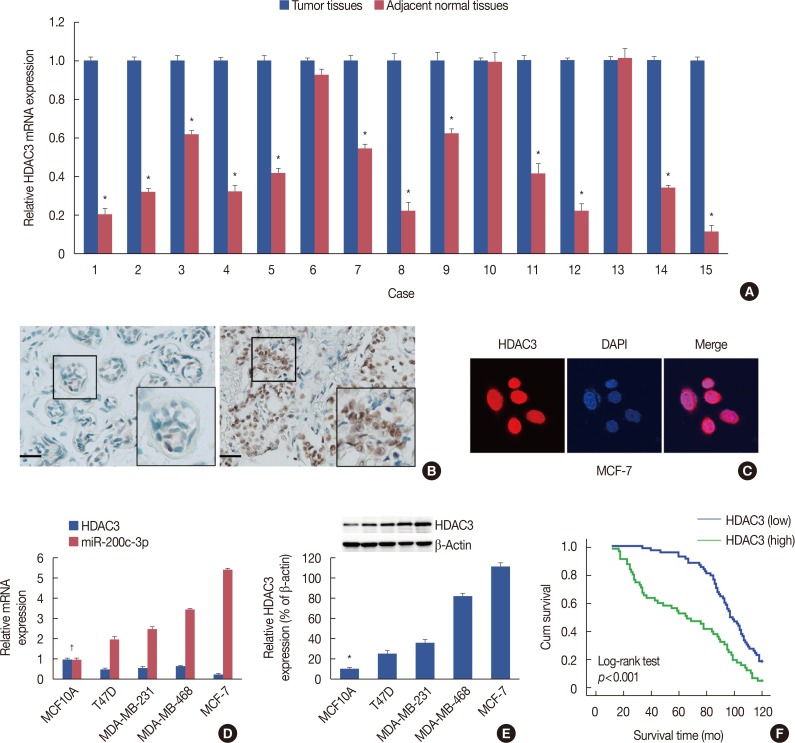 | Figure 1Histone deacetylases-3 (HDAC3) expression in breast cancer. (A) Real-time reverse transcription polymerase chain reaction (RT-PCR) was performed to analyze of messenger RNA (mRNA) expression of HDAC3 in breast cancer tissue of 15 fresh specimens and their paired normal samples. (B) Representative figures of HDAC3 protein expression in adjacent normal tissue (left) and breast cancer tissue (right) using immunohistochemical staining (immunohistochemistry for HDAC3, ×200). (C) Immunofluorescence showed that HDAC3 expression was mainly localized in the cell nucleus in MCF7 cell line. (D) RT-PCR was employed to analyze of mRNA expression of HDAC3 in breast cancer cell lines and normal epithelial breast MCF10A cells. (E) Western blotting analysis of HDAC3 protein expression in breast cancer cell lines and normal epithelial breast MCF10A cells. (F) Association between HDAC3 expression and the prognosis of breast cancer patients.DAPI=4′,6-diamidino-2-phenylindole. *p<0.05; †p<0.05 vs. MCF10A.
|
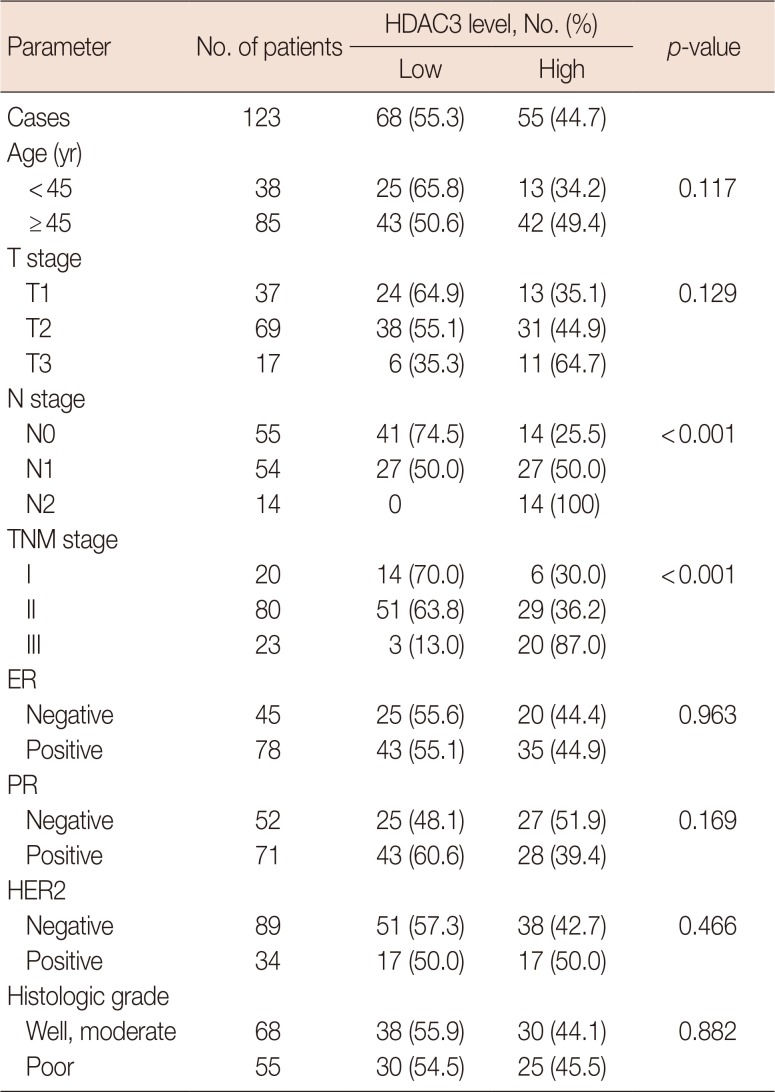
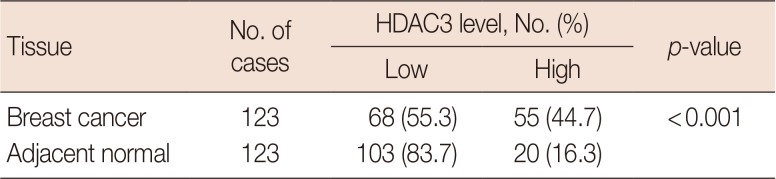
Correlation between HDAC3 expression and clinicopathological characteristics of breast cancer
Table 2
Associations between HDAC3 expression and clinicopathological characteristics in breast cancer
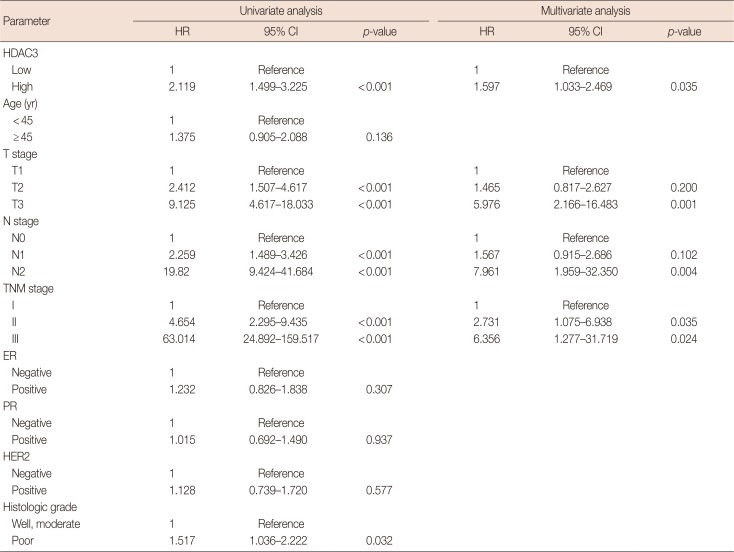
Association between HDAC3 expression and the prognosis of breast cancer patients
Table 3
Summary of univariate and multivariate Cox regression analysis of overall survival duration in all breast cancer patients (n=123)
Knockdown of HDAC3 inhibited breast cancer cell proliferation through suppression of aerobic glycolysis
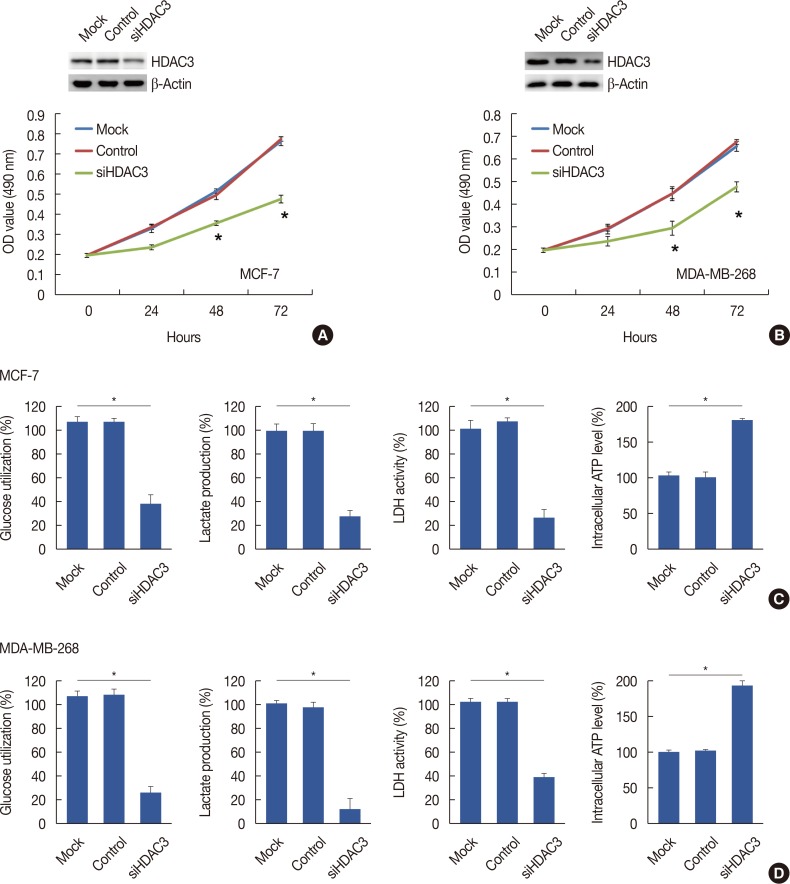 | Figure 2The decrease of cell proliferation and aeroobic glycolysis in response to histone deacetylases-3 (HDAC3) knockdown. The proliferation curve of cells after transfected with control vector or HDAC3 small interfering RNA (siHDAC3) in MCF7 (A) and MDA-MB-268 (B) cell lines, respectively. The glucose utilization, lactate production, lactate dehydrogenase (LDH) activity and intracellular adenosine triphosphate (ATP) level were detected in MCF7 (C) and MDA-MB-268 (D) after transfecting of control vector or siHDAC3.OD=optical density. *p<0.05.
|
Transfection of miR-31 inhibitor can rescue the loss of HDAC3-mediated repression activity in breast cancer cells
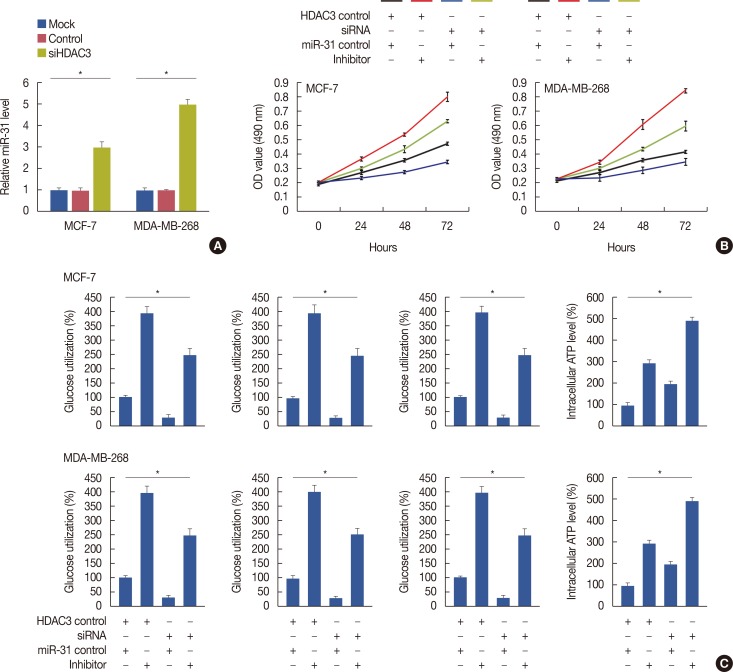 | Figure 3Rescue the loss of histone deacetylases-3 (HDAC3)-mediated repression activity by transfection of microRNA-31 (miR-31) inhibitor in breast cancer. (A) HDAC3 knockdown decreased miR-31 expression. (B) Transfection of miR-31 inhibitor can rescue the loss of HDAC3-mediated reduced cell proliferation in breast cancer. (C) Transfection of miR-31 inhibitor can rescue the loss of HDAC3-mediated decreased lactate dehydrogenase (LDH) activity, glucose utilization, and lactate production and increased intracellular adenosine triphosphate (ATP) level.OD=optical density; siHDAC3=HDAC3 small interfering RNA; siRNA=small interfering RNA. *p<0.05.
|
Overexpression of HDAC3 promoted breast cell proliferation through the promotion of aerobic glycolysis
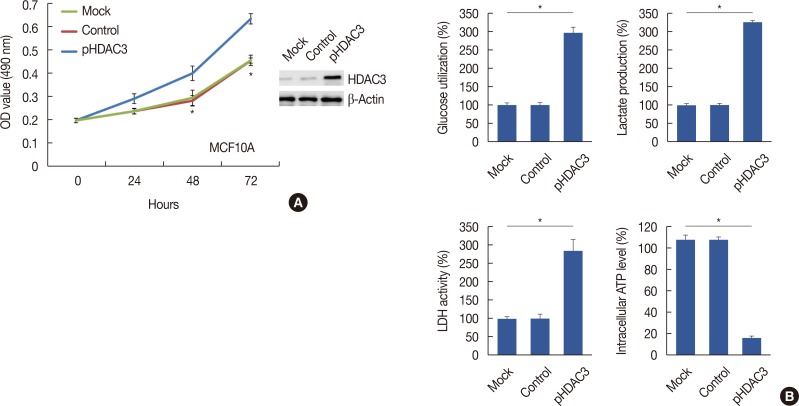 | Figure 4The promotion of cell proliferation and aeroobic glycolysis after histone deacetylases-3 (HDAC3) overexpression. (A) HDAC3 upregulation promoted MCF10A cell proliferation. (B) The glucose utilization, lactate production, lactate dehydrogenase (LDH) activity and intracellular adenosine triphosphate (ATP) level were detected in MCF10A after transfecting of control vector or HDAC3 overexpression (pHDAC3).OD=optical density. *p<0.05.
|
HDAC3 regulated the gene expression of related metabolic enzymes
 | Figure 5Regulation of metabolic enzymes related gene expression by histone deacetylases-3 (HDAC3) level. Inhibition of HDAC3 expression decreased glucose transporter 1 (GLUT-1), GLUT-4, lactate dehydrogenase A (LDHA) and lactate dehydrogenase B (LDHB) expression in MCF7 (A) and MDA-MB-268 (B) cell lines, respectively. (C) Induction of HDAC3 expression increased GLUT-1, GLUT-4, LDHA, and LDHB expression in MCF10A cell.siHDAC3=HDAC3 small interfering RNA; pHDAC3=HDAC3 overexpression. *p<0.05.
|
HDAC3 regulated miR-31 transcriptional activity
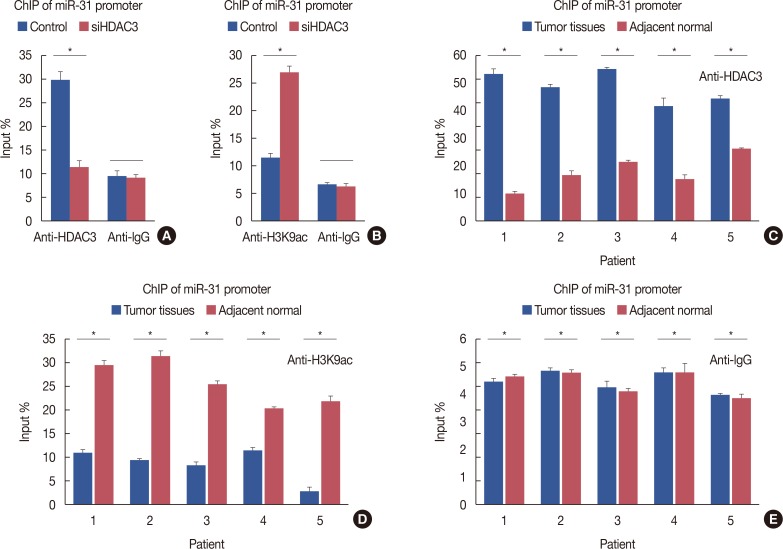 | Figure 6Regulation of microRNA-31(miR-31) transcriptional activity by histone deacetylases-3 (HDAC3). (A) Compared to MCF-7-conHDAC3, MCF-7-siHDAC3 cells exhibited a reduced HDAC3 recruitment to miR-31 promoter. (B) Knockdown of HDAC3 increased histone H3 acetylation at lysine K9 (H3K9ac) levels at the miR-31 promoter. (C) Compared with normal tissues, HDAC3 recruitment to miR-31 promoter was increased in breast cancer tissues. (D) Compared with normal tissues, H3K9ac recruitment to miR-31 promoter was decreased. (E) Normal human IgG was used as a negative control.ChIP=chromatin immunoprecipitation; conHDAC3=HDAC3 control; siHDAC3=HDAC3 small interfering RNA. *p<0.05.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download