Abstract
Tumor suppressor gene O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation has been reported in melanoma. However, the clinical and prognostic significance of MGMT promoter methylation in patients with melanoma remained to be determined. A systematic search was performed to identify eligible papers published. The overall odds ratios (ORs) or hazard ratios and their 95% confidence intervals were calculated. Final 12 eligible publications involving Caucasian population were performed in this study, including 1,071 metastatic melanoma patients, 154 primary melanoma patients, and 211 normal controls. MGMT promoter methylation was significantly higher in primary or metastatic melanoma than in normal controls (p<0.05). No difference of MGMT promoter methylation was found in primary and metastatic melanoma (p=0.432). When metastatic melanoma was compared to normal controls, subgroup analysis showed the correlation between MGMT promoter methylation and different sample materials (tissue: OR=7.01, p<0.001 and blood: OR=12.04, p=0.005). MGMT promoter methylation was not associated with response to drug therapy and the prognosis in overall survival and progression-free survival for multivariate analysis. Our results show that MGMT promoter methylation may be correlated with the increased risk of primary or metastatic melanoma. Based on blood samples, MGMT promoter methylation may become a noninvasive biomarker for the detection of metastatic melanoma. Further additional clinical studies are necessary.
Increasing evidence suggests genetic and epigenetic events, and environmental factor contribute to the development of melanoma123. DNA methylation, a frequent epigenetic modification, is closely associated with the tumorigenesis, progression and prognosis of many types of human cancer45. Mapped to human chromosome 10q26, O6-methylguanine-DNA methyltransferase (MGMT) belongs to a DNA repair gene and counteracts the chemotherapeutic effects of alkylating agent, which protects tumor cells67. MGMT gene within the promoter region is frequently methylated in various cancers, including primary or metastatic melanoma589. However, there are some conflicting and different data about MGMT promoter methylation in patients with melanoma. For example, Marini et al.10 reported that MGMT promoter methylation had a different frequency in primary and metastatic melanoma (64% vs. 15%). Hoon et al.11 reported that MGMT gene had a different promoter methylation level in primary and metastatic melanoma (10% vs. 34%).
Therefore, the present meta-analysis was first conducted to analyze the relationship of MGMT promoter methylation between primary or metastatic melanoma and normal controls. In addition, we analyzed the correlation between MGMT promoter methylation and response to drug therapy in melanoma. Finally, we evaluated the prognostic effect of MGMT promoter methylation in melanoma patients with multivariate analysis.
We systematically searched a range of online digital databases to identify eligible articles published in English up until May 9, 2017, including PubMed, Cochrane Library, Embase and EBSCO. We used the following key words and text word strategy in this meta-analysis: (O6-methylguanine-DNA methyltransferase OR MGMT) AND (methylation OR hypermethylation OR methylated OR epigene*) AND (melanoma OR melanotic cancer OR black cancer). Additionally, a manual reference search for the included articles was also conducted to get other potential publications.
The eligible articles had to satisfy the following inclusion criteria: 1) the patients were clinically diagnosed with primary or metastatic melanoma; 2) studies involved MGMT promoter methylation in melanoma; 3) studies reported sufficient data to evaluate the association between MGMT promoter methylation and melanoma (primary or metastatic melanoma vs. normal controls and metastatic melanoma vs. primary melanoma); 4) studies reported sufficient data to assess the relationship of MGMT promoter methylation clinical features and the prognostic role using multivariate analysis if possible. The study with the most complete information was selected to exclude duplicated publications in the present meta-analysis when authors published more than one article using the overlapping sample data.
Two authors (F.Q. and Z.Y.) independently scanned the eligible publications and extracted the following data. Any disagreements regarding the data were discussed by all authors. The following information was collected from the included studies: the first author's last name, year of publication, mean or median age, tumor stage, treatment information of the patients, country, ethnicity, detection method of MGMT promoter methylation, methylation frequency, the number of cases and controls, treatment response (responder vs. non-responder group), and clinical prognostic information for multivariate analysis.
Stata software (ver. 12.0; Stata Corporation, College Station, TX, USA) was used for the pooled analysis. The pooled odds ratios (ORs) and the corresponding 95% confidence intervals (CIs) were calculated to evaluate the correlation between MGMT promoter methylation and melanoma. In addition, the overall hazard ratios (HRs) and their 95% CIs were also calculated to assess the prognostic role of MGMT promoter methylation in melanoma for multivariate analysis if possible. The Cochran's Q test was used to examine the heterogeneity in the meta-analysis12. Data analysis was performed using the random-effects model. A p-value of <0.1 stands for substantial heterogeneity when heterogeneity was measured1314. Subgroup analyses were conducted to analyze the different association among subgroups.
Through the detailed steps of the systematic literature search, according to the above selection standards (Fig. 1), we finally selected 12 eligible publications in this meta-analysis8910111516171819202122. Articles published in English from 2004 to 2015 included a total of 1,071 patients with metastatic melanoma, 154 patients with primary melanoma, and 211 normal controls. All studies were conducted in Caucasian population. The baseline characteristics of the eligible publications are listed in Table 1.
The data from four studies included 66 patients with primary melanoma and 54 normal controls, as shown in Fig. 2, the pooled OR was 7.49 (95% CI=1.13~49.64, p=0.037), which indicted that MGMT promoter methylation was correlated with an increased risk of primary melanoma.
The data from seven studies consisted of 301 patients with metastatic melanoma and 211 normal controls, as shown in Fig. 2 (OR=7.84, 95% CI=3.58~17.17, p<0.001), that demonstrated that a significant association was found between MGMT promoter methylation and metastatic melanoma.
When 631 patients with metastatic melanoma were compared to 154 patients with primary melanoma, the result from six studies demonstrated that no correlation of MGMT promoter methylation was observed between metastatic and primary melanoma (Fig. 2) (OR=1.63, 95% CI=0.48~5.50, p=0.432).
According to sample type (tissue and blood) and detection method (methylation specific polymerase chain reaction [MSP] and quantitative methylation-specific polymerase chain reaction [QMSP]), we performed subgroup analyses to find the difference among different subgroups.
Subgroup analysis of sample type showed that MGMT promoter methylation was correlated with metastatic melanoma in tissue and blood samples (tissue: OR=7.01, 95% CI=2.90~16.90, p<0.001 and blood: OR=12.04, 95% CI=2.15~67.42, p=0.005) (Fig. 3).
Subgroup analysis by detection method demonstrated that MGMT promoter methylation was associated with metastatic melanoma in the MSP method (OR=8.85, 95% CI=3.83~20.49, p<0.001), but not in the QMSP method (OR=3.37, 95% CI=0.37~30.64, p=0.281) (Fig. 4).
We performed subgroup analysis by therapeutic information (yes vs. not clear) to evaluate the influence of MGMT promoter methylation if patients received drug therapy (heterogeneity, p=0.004). The result showed that no correlation was observed between MGMT promoter methylation and therapeutic information in metastatic vs. primary melanoma (all p>0.1; data not shown).
The pooled OR from six studies including 439 patients with melanoma receiving therapy demonstrated that no relationship was found between MGMT promoter methylation and response to drug therapy (OR=1.82, 95% CI=0.74~4.43, p=0.191) (Fig. 5).
One study involving 122 patients with melanoma receiving therapy reported that MGMT promoter methylation was not associated with overall survival (OS) (HR=0.955, 95% CI=0.63~1.45) and progression-free survival (PFS) (HR=0.91, 95% CI=0.61~1.36) for multivariate analysis9. Another study involving 130 patients with metastatic melanoma receiving therapy reported that MGMT promoter methylation was notably associated with PFS (HR=2.17, 95% CI=1.31~3.57) in multivariate analysis15. While the pooled HR from two studies showed no correlation between MGMT promoter methylation and PFS in multivariate analysis (HR=1.39, 95% CI=0.59~3.25).
DNA methylation of tumor suppressor genes (TSGs) within the promoter region is a major molecular mechanism of epigenomic regulation, which results in the dysfunction or loss of the related gene expression and may play a key role in cancer development2324. MGMT has been identified as a key TSG25, loss of MGMT expression through promoter methylation has been reported in melanoma1115. However, the clinical effect of MGMT promoter methylation remains unclear in melanoma. We performed this meta-analysis to estimate whether MGMT promoter methylation provided valuable insight as an epigenetic biomarker for patients with melanoma.
Our meta-analysis included all eligible articles, which were performed in Caucasian population. The results shown that MGMT promoter methylation was notably higher in primary or metastatic melanoma than in normal controls, which suggested that MGMT promoter methylation may be correlated with the development of melanoma. Additionally, when metastatic melanoma was compared to primary melanoma, MGMT promoter methylation was not found to be statistically significantly higher in metastatic melanoma than in primary melanoma (p=0.432).
Further subgroup analyses were carried out based on the methylation detection method (MSP and QMSP) and sample type (tissue and blood) in the comparison of metastatic melanoma and normal controls. The result showed that the correlation between MGMT promoter methylation and metastatic melanoma was associated with sample type, and MGMT promoter methylation had a higher OR value in the blood (OR=12.04, p=0.005) than in the tissue (OR=7.01, p<0.001), indicating that detection of MGMT promoter methylation may become a potential biomarker using blood samples for metastatic melanoma. Subgroup analysis of testing method showed that MGMT promoter methylation was linked to metastatic melanoma in the MSP subgroup (OR=8.85, p<0.001), but not in the QMSP subgroup (p=0.281), which suggested that MSP may be a sensitive method. However, the subgroup results should be carefully considered as only small sample sizes (blood: 94 cases vs. 96 controls, tissue: 207 cases vs. 115 controls, MSP: 256 cases vs. 185 controls, QMSP: 45 cases vs. 26 controls) were analyzed in this meta-analysis.
MGMT promoter methylation was significantly correlated with response to therapy among two studies1516, but the remaining four studies demonstrated no association between MGMT promoter methylation and response to therapy.891822. Our results involving 439 patients with melanoma from above 6 studies showed that MGMT promoter methylation was not linked to response to therapy (p=0.191). Finally, we evaluated analyzed whether MGMT promoter methylation was associated with the prognosis of melanoma patients for multivariate analysis, promoter methylation of the MGMT gene was not linked to the prognosis in OS among one study9. The result from two studies indicated that MGMT promoter methylation was not correlated with PFS in multivariate analysis. Additional clinical studies with larger sample sizes are needed to further confirm the prognostic significance using multivariate analysis.
Some potential limitations should be stated in this meta-analysis. First, our study only included Caucasian population, such as other study populations, such as Asian and African populations, were not included. Second, only one or two studies reported the prognostic information of MGMT promoter methylation using multivariate analysis. Third, the sample sizes of subgroup analyses were smaller, more studies with large population are essential in the future, especially in blood samples. Finally, in order to compare “future” Asian data, additional studies with the detailed clinical subtype of melanoma (such as acral lentiginous melanoma, superficial spreading melanomas, nodular melanomas, etc.) are needed.
In conclusion, the current findings indicate that MGMT promoter methylation in primary or metastatic melanoma has a higher level than in normal controls. MGMT promoter methylation had a similar frequency in metastatic and primary melanoma, and it may not be correlated with response to drug therapy and the prognosis in OS and PFS. MGMT promoter methylation may be a potential biomarker using blood samples for metastatic melanoma. More prospective studies using large sample sizes are essential to further validate our findings in the future.
References
1. Leucci E, Coe EA, Marine JC, Vance KW. The emerging role of long non-coding RNAs in cutaneous melanoma. Pigment Cell Melanoma Res. 2016; 29:619–626. PMID: 27606977.

2. Berwick M, Buller DB, Cust A, Gallagher R, Lee TK, Meyskens F, et al. Melanoma epidemiology and prevention. Cancer Treat Res. 2016; 167:17–49. PMID: 26601858.

3. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015; 10:103–121. PMID: 25587943.

4. Ye M, Huang T, Ni C, Yang P, Chen S. Diagnostic capacity of RASSF1A promoter methylation as a biomarker in tissue, brushing, and blood samples of nasopharyngeal carcinoma. EBioMedicine. 2017; 18:32–40. PMID: 28396012.

5. Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med (Zagreb). 2015; 25:161–176. PMID: 26110029.
6. Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002; 20:2388–2399. PMID: 11981013.

7. Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000; 343:1350–1354. PMID: 11070098.
8. Beasley GM, Speicher P, Augustine CK, Dolber PC, Peterson BL, Sharma K, et al. A multicenter phase I dose escalation trial to evaluate safety and tolerability of intra-arterial temozolomide for patients with advanced extremity melanoma using normothermic isolated limb infusion. Ann Surg Oncol. 2015; 22:287–294. PMID: 25145500.

9. Hassel JC, Sucker A, Edler L, Kurzen H, Moll I, Stresemann C, et al. MGMT gene promoter methylation correlates with tolerance of temozolomide treatment in melanoma but not with clinical outcome. Br J Cancer. 2010; 103:820–826. PMID: 20736948.

10. Marini A, Mirmohammadsadegh A, Nambiar S, Gustrau A, Ruzicka T, Hengge UR. Epigenetic inactivation of tumor suppressor genes in serum of patients with cutaneous melanoma. J Invest Dermatol. 2006; 126:422–431. PMID: 16374457.

11. Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004; 23:4014–4022. author reply 933. PMID: 15064737.

12. Coory MD. Comment on: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010; 39:932. PMID: 19349478.

13. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003; 327:557–560. PMID: 12958120.

14. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997; 127:820–826. PMID: 9382404.

15. Tuominen R, Jewell R, van den, Wolter P, Stierner U, Lindholm C, et al. MGMT promoter methylation is associated with temozolomide response and prolonged progression-free survival in disseminated cutaneous melanoma. Int J Cancer. 2015; 136:2844–2853. PMID: 25400033.

16. Schraml P, von Teichman A, Mihic-Probst D, Simcock M, Ochsenbein A, Dummer R, et al. Predictive value of the MGMT promoter methylation status in metastatic melanoma patients receiving first-line temozolomide plus bevacizumab in the trial SAKK 50/07. Oncol Rep. 2012; 28:654–658. PMID: 22614944.

17. Cesinaro AM, Sartori G, Migaldi M, Schirosi L, Pellacani G, Collina G, et al. Prognostic significance of MGMT gene promoter methylation in differently treated metastatic melanomas. Pathology. 2012; 44:313–317. PMID: 22544212.

18. Busch C, Geisler J, Lillehaug JR, Lønning PE. MGMT expression levels predict disease stabilisation, progression-free and overall survival in patients with advanced melanomas treated with DTIC. Eur J Cancer. 2010; 46:2127–2133. PMID: 20541396.

19. Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res. 2008; 21:545–558. PMID: 18627528.

20. Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch WCh, Dammann R, et al. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol. 2007; 22:1005–1015. PMID: 17523078.
21. Kohonen-Corish MR, Cooper WA, Saab J, Thompson JF, Trent RJ, Millward MJ. Promoter hypermethylation of the O(6)-methylguanine DNA methyltransferase gene and microsatellite instability in metastatic melanoma. J Invest Dermatol. 2006; 126:167–171. PMID: 16417233.

22. Mori T, O'Day SJ, Umetani N, Martinez SR, Kitago M, Koyanagi K, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005; 23:9351–9358. PMID: 16361635.

23. Ye M, Huang T, Ying Y, Li J, Yang P, Ni C, et al. Detection of 14-3-3 sigma (σ) promoter methylation as a noninvasive biomarker using blood samples for breast cancer diagnosis. Oncotarget. 2017; 8:9230–9242. PMID: 27999208.

24. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054. PMID: 14627790.

25. Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003; 23:206–215. PMID: 12482974.

Fig. 2
Forest plot of the association between O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation and melanoma. OR: odds ratio, CI: confidence interval.
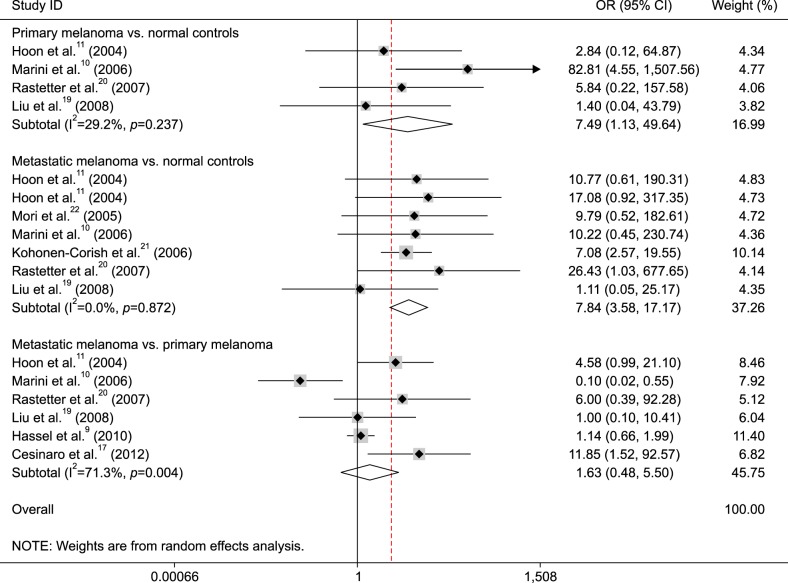
Fig. 3
Subgroup analysis of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation by sample type in metastatic melanoma vs. normal controls. OR: odds ratio, CI: confidence interval.
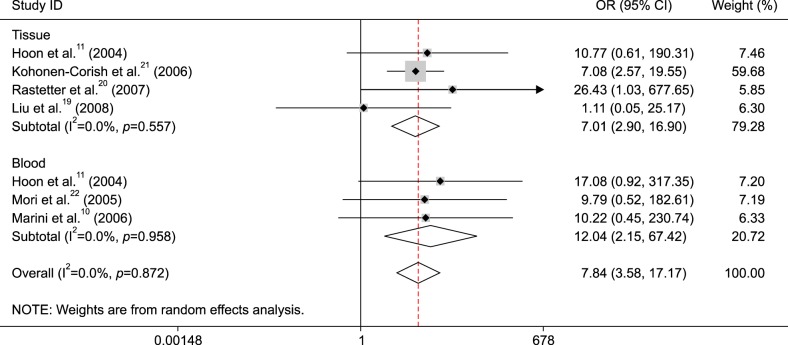
Fig. 4
Subgroup analysis of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation by detection method in metastatic melanoma vs. normal controls. OR: odds ratio, CI: confidence interval, MSP: methylation specific polymerase chain reaction, QMSP: quantitative methylation-specific polymerase chain reaction.
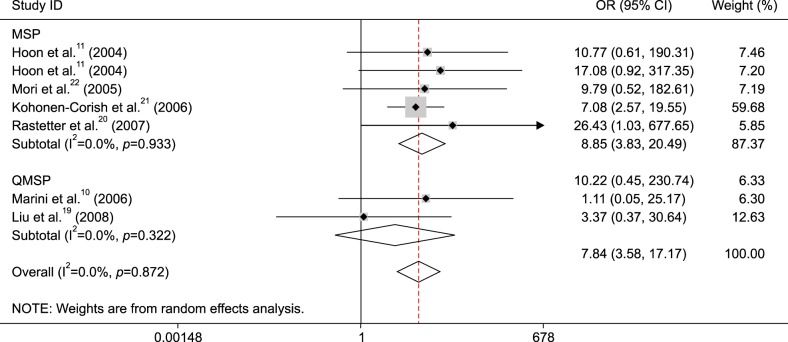
Fig. 5
Forest plot of the association between O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation and response to therapy in melanoma. OR: odds ratio, CI: confidence interval.
Table 1
General characteristics of the eligible publications
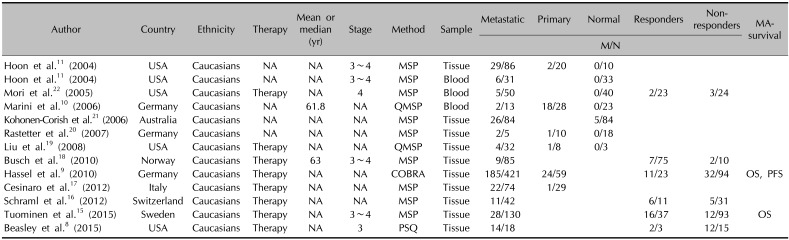
| Author | Country | Ethnicity | Therapy | Mean or median (yr) | Stage | Method | Sample | Metastatic | Primary | Normal | Responders | Non-responders | MA-survival |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M/N | |||||||||||||
| Hoon et al.11 (2004) | USA | Caucasians | NA | NA | 3~4 | MSP | Tissue | 29/86 | 2/20 | 0/10 | |||
| Hoon et al.11 (2004) | USA | Caucasians | NA | NA | 3~4 | MSP | Blood | 6/31 | 0/33 | ||||
| Mori et al.22 (2005) | USA | Caucasians | Therapy | NA | 4 | MSP | Blood | 5/50 | 0/40 | 2/23 | 3/24 | ||
| Marini et al.10 (2006) | Germany | Caucasians | NA | 61.8 | NA | QMSP | Blood | 2/13 | 18/28 | 0/23 | |||
| Kohonen-Corish et al.21 (2006) | Australia | Caucasians | NA | NA | NA | MSP | Tissue | 26/84 | 5/84 | ||||
| Rastetter et al.20 (2007) | Germany | Caucasians | NA | NA | NA | MSP | Tissue | 2/5 | 1/10 | 0/18 | |||
| Liu et al.19 (2008) | USA | Caucasians | Therapy | NA | NA | QMSP | Tissue | 4/32 | 1/8 | 0/3 | |||
| Busch et al.18 (2010) | Norway | Caucasians | Therapy | 63 | 3~4 | MSP | Tissue | 9/85 | 7/75 | 2/10 | |||
| Hassel et al.9 (2010) | Germany | Caucasians | Therapy | NA | NA | COBRA | Tissue | 185/421 | 24/59 | 11/23 | 32/94 | OS, PFS | |
| Cesinaro et al.17 (2012) | Italy | Caucasians | Therapy | NA | NA | MSP | Tissue | 22/74 | 1/29 | ||||
| Schraml et al.16 (2012) | Switzerland | Caucasians | Therapy | NA | NA | MSP | Tissue | 11/42 | 6/11 | 5/31 | |||
| Tuominen et al.15 (2015) | Sweden | Caucasians | Therapy | NA | 3~4 | MSP | Tissue | 28/130 | 16/37 | 12/93 | OS | ||
| Beasley et al.8 (2015) | USA | Caucasians | Therapy | NA | 3 | PSQ | Tissue | 14/18 | 2/3 | 12/15 | |||
M: methylation, N: study population, MA: multivariate analysis, NA: not applicable, MSP: methylation specific polymerase chain reaction, QMSP: quantitative methylation-specific polymerase chain reaction, COBRA: combined bisulphite restriction analysis, OS: overall survival, PFS: progression-free survival, PSQ: pyrosequencing.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download