Abstract
Purpose
Dysregulation of the Wnt pathway is a crucial step in the tumorigenesis of colorectal cancer (CRC). This study aimed to determine whether DNA methylation of Wnt pathway genes helps predict treatment response and survival in patients with metastatic or recurrent CRC.
Materials and Methods
We retrospectively collected primary tumor tissues from 194 patients with metastatic or recurrent CRC. Pyrosequencing was used to examine the methylation of 10 CpG island loci in DNA extracted from formalin-fixed paraffin-embedded specimens. To elucidate the predictive role of DNA methylation markers, Kaplan-Meier survival estimation and Cox regression were performed for progression-free survival and overall survival (OS).
Results
The methylation frequencies of the 10 genes analyzed (p16, p14, MINT1, MINT2, MINT31, hMLH1, DKK3, WNT5A, AXIN2, and TFAP2E) were 47.9%, 10.8%, 21.1%, 16.0%, 20.6%, 0.5%, 53.1%, 32.0%, 2.6%, and 2.1%, respectively. We divided patients into three groups based on the number of methylated genes (group 1, no methylation n=38; group 2, 1–2 methylations n=92; group 3, 3 or more methylations n=64). Among patients treated with palliative chemotherapy (n=167), median OSs of groups 1, 2, and 3 were 39.1, 39.7, and 29.1 months, respectively (log rank p=0.013). After adjustment, number of methylations was identified as an independent poor prognostic factor (0–2 methylated vs. ≥3 methylated: hazard ratio, 1.72; 95% confidence interval, 1.16–2.56, p=0.007).
Colorectal cancer (CRC) is a major public health problem, being the third most commonly diagnosed cancer and the fourth cause of cancer death worldwide.1 In Korea, CRC was predicted to be the third leading cause of cancer death in 2017.2 Although the survival rate of CRC has improved gradually, owing to early screening programs, new surgical techniques, and the development of more-effective systemic therapies, most patients with metastatic or recurrent CRC face death, with a median survival time of 30 months.3 To develop future treatment strategies and prolong the survival of CRC patients, it is crucial to identify the molecular characteristics of CRC, either genomic or epigenomic.
CpG island methylator phenotype (CIMP) is a distinct subtype of various cancers, characterized by an increased frequency of aberrant promoter hypermethylation at specific loci. In cancers with CIMP, aberrant methylation of cytosine nucleotides within these CpG islands can lead to silencing of tumorsuppressor genes, which in turn promotes cancer development. CIMP is reproducibly observed in approximately 15% of CRC cases, and the subgroup defined as CIMP is characterized with high rates of KRAS or BRAF mutation.456 A recent meta-analysis showed that CRC patients with CIMP have a worse prognosis than those without CIMP.7 The CIMP subtype is more enriched in the right side colon when compared with the left side colon, thereby conferring a prognostic impact on CRC.78
Aberrant activation of the Wnt signaling pathway is one of the first events in CRC carcinogenesis. Genomic and epigenomic abnormalities in genes regulating the Wnt pathway, such as APC and AXIN2, are observed in most CRC patients. A decrease in the inhibitory proteins of the Wnt pathway can promote the proliferation and metastasis of CRC, and may be a determinant of a patient's prognosis. DKK 1-3, WNT5A, and AXIN2 are negative regulators of the Wnt pathway, and they are frequently repressed by promotor methylation in CRC.910
AXIN2, one of the genes that predicts high recurrence of surgically resected CRC, is derived from cancer stem cells and has been incorporated into methylation signatures.11 A German group reported that hypermethylation of TFAP2E results in downregulation of DKK4 and is a predictor of chemotherapy resistance in CRC.12 CRC with loss of WNT5A expression and hypermethylation of WNT5A was reported to be associated with a poor prognosis in a previous study.13
The aim of this study was to evaluate the prognostic value of the methylation status of four Wnt pathway genes (DKK3, WNT5A, AXIN2, and TFAP2E) in metastatic or recurrent CRC patients. In addition, we explored the prognostic impact of the methylation status of an additional six genes from the classic CIMP panel.
We included 236 patients with metastatic or recurrent CRC whose primary or metastatic tumor specimens were sufficient for evaluating methylation status by pyrosequencing. All patients were treated with first-line chemotherapeutics at Severance Hospital, Yonsei University from June 1999 to June 2010. Demographic and clinical information was obtained from the electronic medical records of Severance Hospital, and survival data were retrieved from the tumor registry at the Yonsei Cancer Center. Exclusion criteria comprised co-existing malignancies (except for non-melanoma skin cancer or in situ cervical cancer), cancer other than adenocarcinoma, and insufficient amount of formalin-fixed paraffin-embedded (FFPE) tumor tissue for DNA extraction. This study was approved by the Institutional Review Board (IRB) at Yonsei University Severance Hospital, Seoul, Korea (IRB Number 4-2013-0134).
Genomic DNA from FFPE tissues was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA). Extracted tumor DNA was used to evaluate the mutation status of KRAS at codons 12 and 13 and BRAF at codon 600 and the methylation status of six CpG islands from a classic CIMP panel (MINT1, MINT2, MINT31, hMLH1, p16, and p14) and four Wnt pathway-related genes (DKK3, WNT5A, AXIN2, and TFAP2E). The DNA was first treated with bisulfite, using an EZ Methylation kit (Zymo Research, Orange, CA, USA), and then pyrosequencing was performed using the PyroMark Q24 instrument (Qiagen). The detailed protocol for DNA mutation and methylation status evaluation using a pyrosequencer has been described in previous reports.814 The methylation status of CpG island markers was based on a threshold value of 15% (negative, methylation level <15%; positive, methylation level ≥15%). The numbers of CpGs analyzed in the pyrosequencing assay were 2 for AXIN2, DKK3, and TFAP2E and 3 for WNT5A. Given that the concordance in methylation between the adjacent CpGs was very high for all four genes, we used average methylation for our analysis. A CIMP-positive tumor was defined as one in which two or more CIMP markers were methylation positive. Patients were divided into three groups based on the methylation status of the 10 evaluated genes (group 1, 0 methylation; group 2, 1–2 methylations; group 3, 3 or more methylations).
All patients were treated with 5-fluorouracil, leucovorin, oxaliplatin, and irinotecan as single agents or in combination. The dose and schedule of the FOLFOX and FOLFIRI regimens were the same as those described in other studies or guidelines. As a routine clinical protocol, tumor assessment for response to chemotherapy was evaluated every 8 to 12 weeks, using CT scanning or magnetic resonance imaging in accordance with the Response Evaluation Criteria in Solid Tumors (RECIST) ver. 1.1. Overall survival (OS) was calculated from the first day of diagnosis to the day of death or the last follow-up visit. Progression-free survival (PFS) was calculated from the first day of chemotherapy to the time of the first documentation of progression, death from any cause, or the date of last follow-up visit if no events had occurred. Objective response rate (ORR) was calculated as the proportion of complete response (CR) and partial response (PR) by RECIST criteria.
Clinicopathologic variables were used as categorical variables. The difference in categorical variables and ORR was assessed by the chi-square or Fisher's exact test. Survival curves were estimated by the Kaplan-Meier method for OS and PFS, with 95% confidence interval (CI) for the median time to event. The log-rank test was used to compare the distribution of survival between groups in a univariate analysis. Multivariate analyses were carried out using the Cox proportional hazard regression model, including variables with p<0.10 in the univariate analysis. Two-sided p-values of <0.05 were considered to indicate statistical significance. All analyses were performed using SPSS for Windows, version 18.0 (SPSS Inc., Chicago, IL, USA).
A total of 194 patients had sufficient amounts of tumor DNA extract for methylation analysis (Table 1, Supplementary Tables 1 and 2, only online). Approximately two-thirds (n=127) of the patients were male, and half (n=107) had metastatic CRC at the time of initial diagnosis. The primary tumor was located in the right and left side of the colon in 22.2% (n=43) and 77.9% (n=151) of patients, respectively. Microsatellite instability (MSI) status was known in 140 patients, of which five were MSI-high. Based on the classic definition of CIMP, 59 (30.4%) patients were classified as CIMP positive. Genomic tests of KRAS and BRAF and the methylation status of Wnt genes are summarized in Table 2. Mutations in KRAS and BRAF were detected in 64 (33.0%) and 8 (4.1%) patients, respectively. CIMP-positive tumors harbored a significantly higher frequency of KRAS mutation than CIMP-negative tumors did (44.1% in CIMP+ vs. 28.1% in CIMP-; chi-square, p=0.03) (Fig. 1). The frequency of BRAF mutation was also significantly higher in CIMP+ tumors (11.9% in CIMP+ vs. 0.7% in CIMP-; Fisher's exact test, p=0.01) (Fig. 1). Based on methylation status, 38 (19.6%), 92 (47.4%), and 64 (33.0%) patients were divided into groups 1, 2, and 3, respectively. The mean number of methylations was higher in proximal-located CRC tumors (2.63, standard deviation 2.11) than in distal-located CRC tumors (1.91, standard deviation 1.72). The most frequently methylated marker in group 3 was p16 (85.9%; 55 of 64 patients).
Among the 194 patients included, 167 received palliative first-line chemotherapy after diagnosis of metastatic or recurrent CRC. Surgical removal of metastatic lesions was amenable in 39 (23.4%) patients, and they were excluded from the ORR analysis. The 27 patients who did not receive palliative chemotherapy were excluded from both the ORR and survival analyses. More than two-thirds of the patients who received palliative first-line treatment were treated with either FOLFOX or FOLFIRI (n=114, 68.3%). The remaining patients were treated with either molecular targeted agents, such as bevacizumab or cetuximab (n=35, 20.9%), or single agents, such as 5-FU or Capecitabine (n=18, 10.8%). In 128 patients who did not undergo metastasectomy, ORR (CR+PR) was observed in 63 (49.2%), and 10 (7.8%) had only non-measurable lesions according to the RECIST criteria. The number of methylations in a CRC tumor sample was significantly associated with the ORR of first-line chemotherapy (Fig. 2). The ORR was 66.7%, 46.3%, and 43.2% in group 1, 2, and 3, respectively. Only one (4.2%) patient experienced progressive disease (PD) in group 1, whereas nine (13.4%) had PD in groups 2 and 3.
With a median follow-up time of 38 months, 131 (67.5%) of the 194 patients died. However, we only selected the 167 patients who received first-line chemotherapy for our survival analysis. None of the 10 methylation sites was associated with OS in a univariate analysis. CIMP-positive patients exhibited a trend toward shortened survival, when compared with CIMP-negative patients (log-rank, p=0.16) (Fig. 3A). Nevertheless, we found that a higher number of methylations was associated with shorter OS. The median OS was 39.1, 39.7, and 29.1 months in groups 1, 2, and 3, respectively (log-rank, p=0.013) (Fig. 3B). In distal-located primary tumors, there was a statistically significant association between the number of methylations and the OS (n=132, median OS 39.1 months in group 1 vs. 43.0 months in group 2 vs. 31.2 months in group 3; p=0.011); however, this was not observed in proximal-located tumors (n=35, median OS 34.7 months in group 1 vs. 25.5 months in group 2 vs. 29.1 months in group 3; p=0.688).
Group 1 had statistically non-significant longer PFS of first-line chemotherapy than group 2 or 3 did. The median PFS was 9.2, 7.6, and 7.0 months in groups 1, 2, and 3, respectively (log-rank, p=0.198). In the 128 patients who did not undergo metastasectomy, the median PFS was 9.1, 6.4, and 6.7 months, respectively (log-rank, p=0.075). This trend toward improved survival in group 1 was also observed in the 39 patients who underwent metastasectomy: not reached, 49.6 months and 44.4 months in groups 1, 2, and 3, respectively (p=0.429).
Among the variables that were significantly associated with OS in the univariate analyses [age, Eastern Cooperative Oncology Group performance status (ECOG PS), histologic grade, American Joint Committee on Cancer (AJCC) stage at diagnosis, BRAF mutation, resectability at the time of metastasis, objective response of chemotherapy], the number of methylations was independently associated with decreased survival in all patients (OS for 0–2 methylations vs. ≥3 methylations; hazard ratio, 1.72, 95% CI, 1.16–2.56; p=0.007) (Table 3).
In this study, we evaluated the methylation status of DKK3, WNT5A, AXIN2, and TFAP2E in patients with metastatic or recurrent CRC. Although a single methylation of one of the four genes was not associated with the prognosis of our subjects, higher numbers of methylations among the 10 evaluated genes was independently associated with poorer clinical outcome. In chemotherapy-treated CRC patients, OS was significantly longer in groups 1 and 2 than in group 3. In patients with measurable disease, the ORR was significantly higher in group 1 than in group 2 or 3, and the median PFS was numerically higher in group 1 than in group 2 or 3.
We selected four Wnt pathway genes and six classic CIMP panel genes because aberrant genomic or epigenomic changes of the Wnt signaling pathway are crucial in CRC carcinogenesis and progression. Methylation of DKK3 was present in 53.1% of our patients, and was not associated with survival. Similarly, in a previous study by Yu, et al.,15
DKK3 methylation was detected in 67 of 128 (52.3%) patients with CRC tumors, and was not predictive of patient survival. However, they reported that DKK3 methylation status may be of potential use as a prognostic marker in patients with gastric cancer who may benefit from aggressive treatment. Methylation of WNT5A has been detected in 18–54% of CRC tumors in previous studies.81617
WNT5A has been studied as an important functional and prognostic marker in various cancers. In CRC, Rawson, et al.17 evaluated the largest cohort of 1232 patients, and showed that WNT5A was strongly associated with sporadic MSI-high tumors without prognostic implication, in concordance with the present study. These findings may indicate that the role of the Wnt signaling pathway varies between CRC subtypes, depending on the epigenetic event of related genes.
Methylation of AXIN2 and TFAP2E was detected only in 5 (2.6%) and 4 (2.1%) patients, respectively, in the current study. Therefore, the number of patients was too small to analyze the prognostic impact of these genes. There are several possible reasons for why the frequency of methylation in both genes was lower than that reported in other western studies. First, the Asian ethnicity of our study population could have influenced the results. Yoon, et al.18 reported that CRC from Asians have a lower rate of BRAF and KRAS mutations than that from people of African descent and Caucasians. Given that previous studies have reported that methylation of AXIN2 and TFAP2E is correlated with BRAF mutation, the tumors in our study population may have intrinsically lower rates of methylation. Second, the study method used to detect CpG island methylation was different from that used in previous studies. Pyrosequencing is not as sensitive, but is specific, for detecting methylation status, compared to methylation-specific PCR, which has been utilized in many previous studies. However, some proportions of AXIN2 and TFAP2E methylation could be false positives, given that miss-priming of methylation-specific PCR frequently occurs during experiments.19 In addition, low tumor purity or tumor heterogeneity of biopsy specimens from metastatic CRC patients may have contributed to the low frequency of methylation: other studies examined the methylation status in operation specimens from early CRC patients.
Wnt pathway methylation is easy to assess in routine clinical practice, when compared with whole-genome or transcriptome analysis. In addition, the advantage of a DNA methylation marker is its stability and reproducibility, in contrast to RNA-based biomarkers, and actionability with demethylating agents, such as 5-Aza. Accordingly, recently, an international consortium attempted to share large-scale data and enable the classification of CRC into four consensus molecular subtypes (CMSs).20 CMS4 subtype CRC patients exhibit worse prognosis than patients with other subtypes, and show low Wnt activity, which might have resulted from repression of Wnt pathway genes by CpG island methylation. Interestingly, De Sousa E Melo, et al.11 documented that a stem cell-derived gene signature (mesenchymal features with chemotherapeutic resistance and high TGF-β signaling) was closely related to methylation-dependent tuning of the Wnt expression program.
The main limitations of our study are its retrospective nature and the heterogenous treatment protocols. Therefore, our results need to be validated in future prospective studies. Another study limitation is that we did not evaluate the extended RAS mutations (in exons 3 and 4 of KRAS and exons 2, 3, and 4 of NRAS) or the MSI status of all patients. However, this should not have a significant impact on our results because most of our patients did not receive anti-EGFR monoclonal antibody. Lastly, the absence of a consensus panel defining CIMP subgroups is a hurdle to be overcome with comparison with other future studies.
In conclusion, we found that a higher number of methylations among the 10 genes evaluated, comprising four Wnt pathway genes and six classic CIMP panel genes, was independently associated with poorer clinical outcomes in CRC patients. Further validation of these markers in prospective settings and early stage patients will guide CRC patients to more personalized treatment planning and, hopefully, improve clinical outcomes in the future.
ACKNOWLEDGEMENTS
This study was supported by a faculty research grant of Yonsei University College of Medicine for 2010 (6-2010-0022).
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.

2. Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH. Prediction of cancer incidence and mortality in Korea, 2017. Cancer Res Treat. 2017; 49:306–312. PMID: 28301926.

3. Cremolini C, Schirripa M, Antoniotti C, Moretto R, Salvatore L, Masi G, et al. First-line chemotherapy for mCRC—a review and evidence-based algorithm. Nat Rev Clin Oncol. 2015; 12:607–619. PMID: 26215044.

4. Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000; 97:710–715. PMID: 10639144.

5. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009; 361:2449–2460. PMID: 20018966.
6. Sambuudash O, Kim HS, Cho MY. Lack of aberrant methylation in an adjacent area of left-sided colorectal cancer. Yonsei Med J. 2017; 58:749–755. PMID: 28540987.

7. Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol. 2014; 25:2314–2327. PMID: 24718889.

8. Ahn JB, Chung WB, Maeda O, Shin SJ, Kim HS, Chung HC, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011; 117:1847–1854. PMID: 21509761.

9. Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006; 25:4116–4121. PMID: 16491118.

10. Koinuma K, Yamashita Y, Liu W, Hatanaka H, Kurashina K, Wada T, et al. Epigenetic silencing of AXIN2 in colorectal carcinoma with microsatellite instability. Oncogene. 2006; 25:139–146. PMID: 16247484.

11. de Sousa E Melo F, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011; 9:476–485. PMID: 22056143.

12. Ebert MP, Tänzer M, Balluff B, Burgermeister E, Kretzschmar AK, Hughes DJ, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med. 2012; 366:44–53. PMID: 22216841.
13. Dejmek J, Dejmek A, Säfholm A, Sjölander A, Andersson T. Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res. 2005; 65:9142–9146. PMID: 16230369.

14. Kim SH, Park KH, Shin SJ, Lee KY, Kim TI, Kim NK, et al. p16 Hypermethylation and KRAS mutation are independent predictors of cetuximab plus FOLFIRI chemotherapy in patients with metastatic colorectal cancer. Cancer Res Treat. 2016; 48:208–215. PMID: 25943321.
15. Yu J, Tao Q, Cheng YY, Lee KY, Ng SS, Cheung KF, et al. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer. 2009; 115:49–60. PMID: 19051296.
16. Ying J, Li H, Yu J, Ng KM, Poon FF, Wong SC, et al. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/beta-catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008; 14:55–61. PMID: 18172252.
17. Rawson JB, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, et al. Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. Br J Cancer. 2011; 104:1906–1912. PMID: 21587258.

18. Yoon HH, Shi Q, Alberts SR, Goldberg RM, Thibodeau SN, Sargent DJ, et al. Racial differences in BRAF/KRAS mutation rates and survival in stage III colon cancer patients. J Natl Cancer Inst. 2015; 107:djv186. PMID: 26160882.
19. Shaw RJ, Akufo-Tetteh EK, Risk JM, Field JK, Liloglou T. Methylation enrichment pyrosequencing: combining the specificity of MSP with validation by pyrosequencing. Nucleic Acids Res. 2006; 34:e78. PMID: 16807314.

20. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015; 21:1350–1356. PMID: 26457759.
Fig. 1
Distribution of mutations and methylation in our patients. Each bar represents individual mutations (light blue) and methylations (dark blue). CIMP is denoted by an orange bar. CIMP-positive tumors had more frequent KRAS or BRAF mutation than CIMP negative tumors. CIMP, CpG island methylator phenotype.
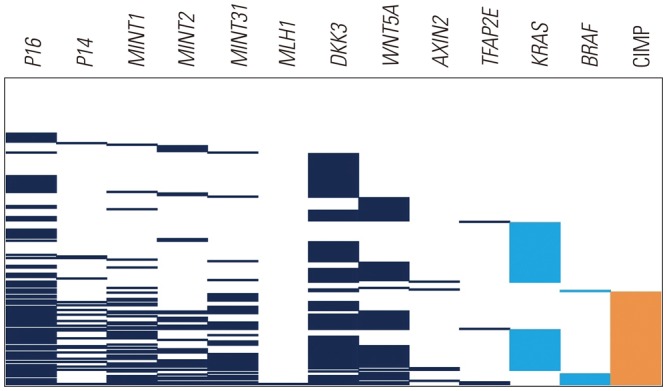
Fig. 2
Bar chart showing the best objective response of first-line chemotherapy in each group. ORR was higher in group 1 (66.7%) than in group 2 (46.3%) or 3 (43.2%). CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; UA, unavailable, ORR, objective response rate.
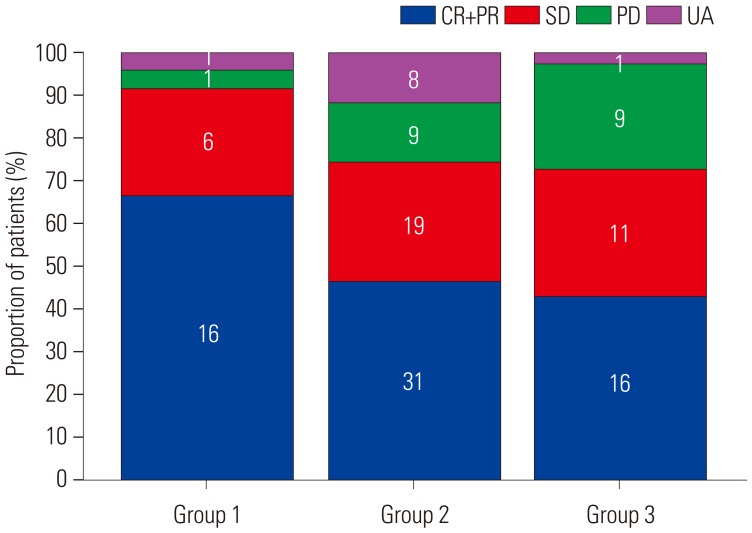
Fig. 3
Overall survival according to CpG island phenotype (A) and the number of methylations (B). CIMP, CpG island methylator phenotype.
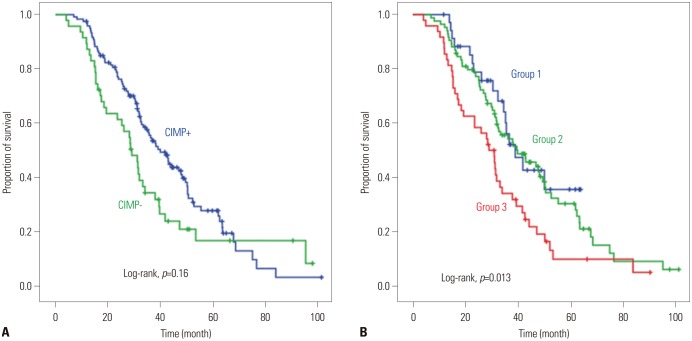
Table 1
Patient Characteristics
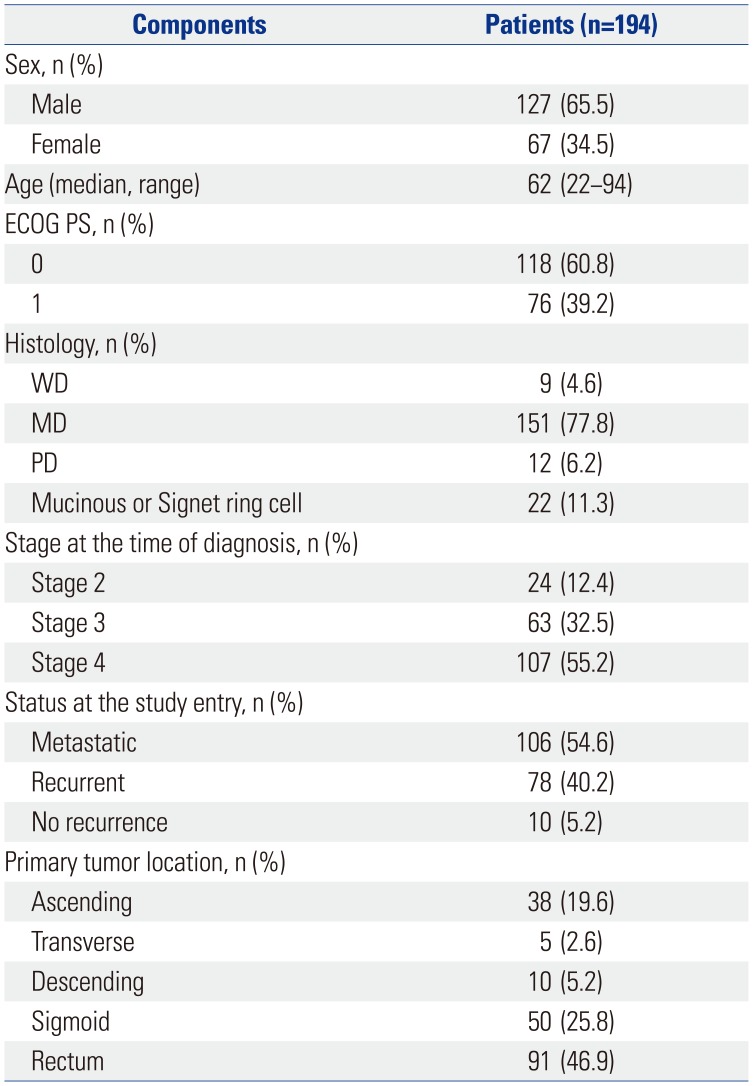
Table 2
Molecular Profile of Tumors
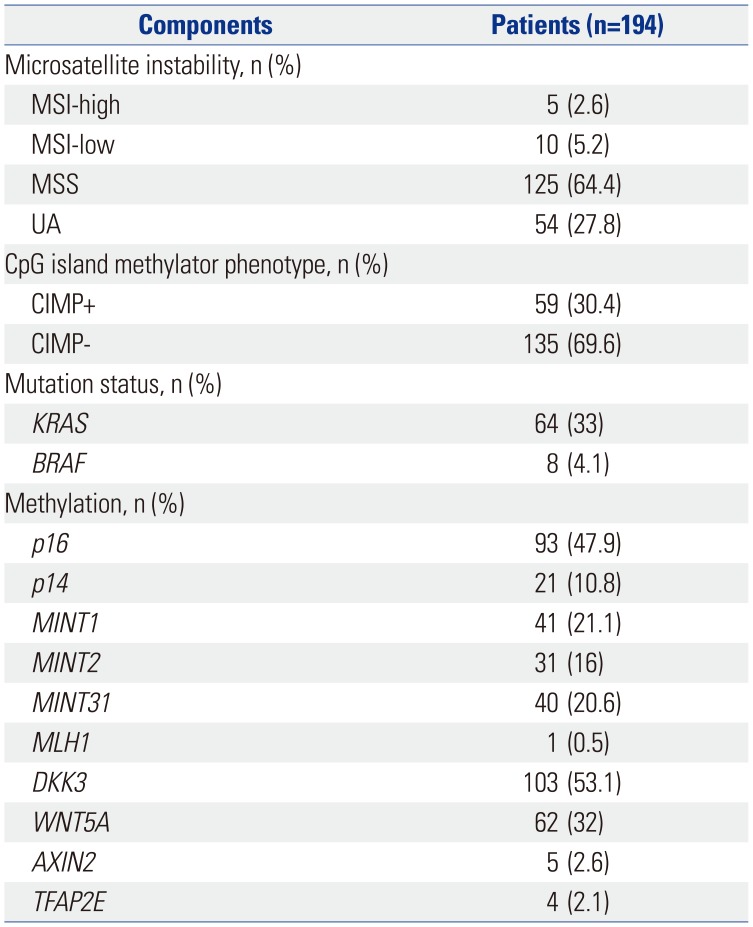
Table 3
Univariate and Multivariate Analyses for Overall Survival
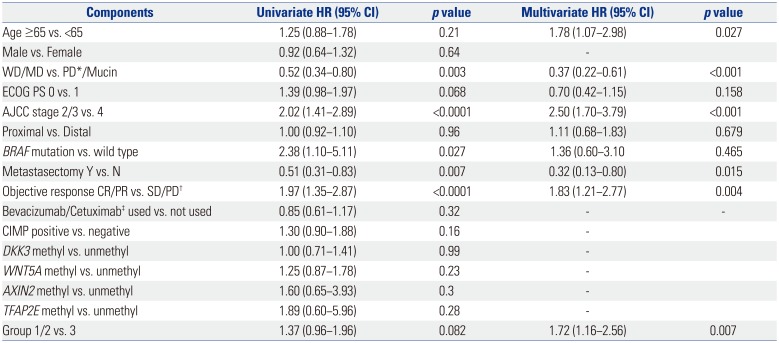
ECOG PS, Eastern Cooperative Oncology Group performance status; WD, well differentiated; MD, moderately differentiated; PD*, poorly differentiated; AJCC, American Joint Committee on Cancer; CR, complete response; PR, partial response; SD, stable disease; PD†, progressive disease; CIMP, CpG island methylator phenotype; group 1, no methylation; group 2, 1–2 methylation; group 3, 3 or more methylation; HR, hazard ratio; CI, confidence interval.
‡Bevacizumab was used in 22 patients and cetuximab was used in 16 patients.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download