Abstract
Two strains of porcine reproductive and respiratory syndrome virus (PRRSV) were isolated in 2006 and 2016 and designated as FZ06A and FZ16A, respectively. Inoculation experiments showed that FZ06A caused 100% morbidity and 60% mortality, while FZ16A caused 100% morbidity without death. By using genomic sequence and phylogenetic analyses, close relationships between a Chinese highly pathogenic PRRSV strain and the FZ06A and FZ16A strains were observed. Based on the achieved results, multiple genomic variations in Nsp2, a unique N-glycosylation site (N33→K33), and a K151 amino acid (AA) substitution for virulence in the GP5 of FZ16A were detected; except the 30 AA deletion in the Nsp2-coding region. Inoculation experiments were conducted and weaker virulence of FZ16A than FZ06A was observed. Based on our results, a 30 AA deletion in the Nsp2-coding region is an unreliable genomic indicator of a high virulence PRRSV strain. The Nsp2 and GP5 differences, in addition to the virulence difference between these two highly pathogenic PRRSV strains, have the potential to be used to establish a basis for further study of PRRSV virulence determinants and to provide data useful in the development of vaccines against this economically devastating disease.
Porcine reproductive and respiratory syndrome virus (PRRSV) belongs to the Arteriviridae family and the genus Nidovirales, which includes viruses with epidemiological relevance such as lactate dehydrogenase-elevating virus of mice, equine arteritis virus, and simian hemorrhagic fever virus [730]. PRRSV was first reported in 1987 in North America [616], and later in Netherlands and other parts of the world [28]. PRRSV has strong pathogenicity with a high transmissible effect. The virus causes morbidity and mortality rates in the range of 50% to 100% and 20% to 100%, respectively [33]. PRRS has resulted in huge economic losses in the livestock industry worldwide. In 2006, highly pathogenic PRRSV (HP-PRRSV) was first identified in China, where it affected vaccinated pigs (in all ages) by producing a high fever and resulted in a high mortality rate. Afterward, it rapidly spread to neighboring countries, in which their swine industries experienced widespread losses [226].
The genome of PRRSV is a single-stranded, positive-sense RNA of approximately 15 kb length that contains a 5′-capped structure and a 3′-polyadenylated tail. At least nine open reading frames (ORF1a, ORF1b, ORF2, ORF3, ORF4, ORF5a, ORF5, ORF6, and ORF7) that are flanked by 5′ and 3′ untranslated regions (UTR) have been identified [121723]. ORF1a and ORF1b encode non-structural proteins (NSPs). ORFs 2 to 7 encode PRRSV structural proteins, including small envelope (E), GP2, GP3, GP4, GP5a, GP5, matrix (M), and nucleocapsid (N) proteins [15]. Based on their genetic diversity, PRRSVs are classified into two genotypes: genotype I (European type) and genotype II (North American type). These two genotypes share only about 55% to 70% nucleotide identity and 50% to 80% amino acid (AA) sequences [4]. Within each genotype, the PRRSV presents extensive genetic variations that reflect the diversity of the strains [1324]. Moreover, the Nsp2 and ORF5 regions of the PRRSV genome display substantial genetic variation. These regions have been used as markers for genetic variability because they are the most variable regions [911]. In recent years, continuing evolution of PRRSV has resulted in the emergence of novel strains with different levels of pathogenicity and virulence [183637383940]. That persistent variation and evolution of PRRSV is an intractable issue in the effective control of PRRS.
In this study, we sequenced two PRRSV strains isolated in 2006 and 2016 and designated them as FZ06A and FZ16A, respectively. Clinical signs, pathological changes, and humoral immune responses of the isolated strains were analyzed to evaluate the difference between the two HP-PRRSV strains and to provide an improved description of the persistent variation and evolution of PRRSV. The aim of this research was to contribute to the development of a constructive and effective database that would be useful in the development of medical vaccines against this economically devastating disease.
All animals used in this research were treated with care and with the approval of the Animal Care and Use Committee of the Chinese Academy of Agricultural Sciences, China (IACUC No. PJ2011-012-03).
In 2006 and 2016, two strains of PRRSV (FZ06A and FZ16A, respectively) were isolated from pigs having high fever, labored breathing, lethargy, and anorexia in Fujian province, South China. In 2006, a pig farm had an outbreak of PRRSV from which a high death rate was observed. Likewise, in 2016, the same pig farm had a high morbidity related to PRRSV but there was no large-scale mortality. Isolated strains of the PRRSVs, as clinical samples, were immersed in sterile phosphate-buffered saline with antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) and the suspension was centrifuged at 10,000 × g for 10 min to extract RNA and isolate the virus. The remaining samples were filtered and kept at −80℃ until needed.
Marc-145 cells were used for virus isolation. The virus sample was filtered (through 0.22 µm filter) and then inoculated to Marc-145 cells. After adsorption for 1 h at 37℃, the supernatant was removed and Dulbecco's modified Eagle's medium (Gibco, USA) with 2% fetal bovine serum (Gibco) was added. Then, the cells were incubated at 37℃ with 5% CO2 and monitored daily for cytopathic effects (CPEs). When CPEs appeared in 80% of the cells, the culture supernatants were harvested and stored at −80℃ until needed.
Viral RNA was extracted by using AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen Biosciences, USA) from the original tissue homogenates and cell culture supernatants, and reverse transcription polymerase chain reaction (RT-PCR) was carried out with a PrimeScript one-step RT-PCR Kit (Takara, China) according to the manufacturer's instructions. Fourteen pairs of specific primers (Table 1) were designed for the amplification and were based on the complete genomic sequences of PRRSV in the GenBank database (National Center for Biotechnology Information, USA). Fourteen overlapped fragments that covered the entire viral genome were amplified by RT-PCR. The PCR conditions involved an initial denaturation step at 94℃ for 2 min, followed by 30 cycles of 94℃ for 30 sec, 52℃ to 55℃ for 30 sec, 72℃ for 2 min, and final extension at 72℃ for 10 min. The amplified products were analyzed by electrophoresis in a 1% agarose gel.
The presence and morphology of the purified virus were evaluated by transmission electron microscopy (TEM) of negatively stained preparations. Briefly, carbon-coated grids were set on a 20 µL sample drop for 10 min and then washed with deionized water. The washed grids were then negatively stained with 1% uranyl acetate at pH 4.5. After removal of the excess stain by using filter paper, the grids were observed under a TEM [22].
The amplified PCR products were purified by using an E.Z.N.A. gel extraction kit (OMEGA; Bio-Tek, USA). The PCR amplicons were then cloned into a pEAZY-blunt-zero vector according to the manufacturer's instructions (TransGen, China) and sequenced. The full-length genomes of the new isolates were assembled according to the sequencing data by using Lasergene 7.2 software (DNASTAR, USA). By utilizing MEGA software (ver. 5.1) [31], the assembled genomes were aligned with reference PRRSV strains in the GenBank database (Table 2). The sequences of the complete genome, as well as those of Nsp2 and ORF5, were compared with other PRRSV isolates. Phylogenetic trees were constructed with MEGA software (ver. 5.1) by using the neighbor-joining method. Bootstrap values were calculated on 1,000 replicates of the alignment.
By using the BioEdit sequence alignment editor software (ver. 7.2.5; Ibis Therapeutics, USA), the nucleotide sequences obtained after completion of genomic sequencing analysis were translated into the predicted AA sequences. For the comparative analysis, representative sequences of PRRSV were also selected from GenBank (GenBank accession numbers: AY150564, AY032626, AY150312, EF075945, EF635006, EF112445, JX087437, EU144079, EU860248, EU187484, and AF331831).
Fifteen 9-week-old, specific-pathogen-free (SPF) pigs, which were negative for antibodies of porcine circovirus type 2 (PCV2) and PRRSV, were randomly divided into three groups (five pigs per group): two challenge groups (groups 1 and 2) and a control group (group 3). The third virus passage (P3) from Marc-145 cells was used in these experiments. The selected pigs in groups 1 and 2 were inoculated intranasally (i.n.) with 2 mL inocula that contained 105 50% tissue culture infective doses (TCID50) of the PRRSV strains FZ06A and FZ16A. The group 3 animals were inoculated i.n. with 2 mL of Marc-145 cell culture supernatant.
After infection of the pigs, clinical signs, including depression, appetite, diarrhea, skin discoloration, coughing, labored and abdominal breathing, and respiratory rate of the animals were monitored twice a day. Rectal temperatures were recorded every day throughout the experiment until completion at 21 days post-inoculation (dpi).
Blood samples were collected at 1, 3, 7, 10, 14, and 21 dpi followed by culturing at 37℃ for 1 h. Subsequently, the blood was centrifuged at 3,500 × g for 5 min and the serum was frozen at −80℃. Serum samples underwent 10-fold serial dilution and were then incubated with Marc-145 cells in 96-well cell culture plates at 37℃ for 2 h. After incubation for 48 h, cells were fixed with 4% paraformaldehyde. The viral titers were determined by an indirect fluorescent antibody (IFA) technique with anti-N protein monoclonal antibody against PRRSV N protein and FITC-labeled goat-anti-mouse IgG (ZSGB-BIO, China). Viral titers were calculated as TCID50 per mL by using the Reed-Muench method.
Serum was collected at 1, 3, 7, 10, 14, and 21 dpi, and presence of PRRSV-specific antibodies was measured by using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (IDEXX Laboratories, USA), according to the manufacturer's instructions. The positive and negative cut-offs of the IDEXX ELISA were set at a sample-to-positive (S/P) ratio of 0.4.
At 21 dpi, all infected and control animals were euthanized and tissue samples of lung, spleen, kidney, liver, and lymph nodes were collected for detection of virus by means of RT-PCR assays. A similar analysis method (RT-PCR) was used to determine the presence of pseudorabies virus (PRV), PCV, and classical swine virus (CSFV) in all tissue samples.
At necropsy, lungs of the investigated animals were fixed in 10% neutral buffered formalin and then analyzed for macroscopic (gross) pathologies. In addition, some of the lungs tissues collected were processed for histopathological examination followed by H&E staining, as described previously [14].
GraphPad Prism 5 software (GraphPad, USA) was used for analysis of the variability among the groups, and data were expressed as mean ± SD values. By using one-way ANOVA and Tukey's t-tests, the results were statistically evaluated. Differences were considered statistically significant when p < 0.05.
The two isolated viruses were tested as PRRSV positive by RT-PCR assays. After three passages on Marc-145 cells, a distinct CPE was observed (Fig. 1). These isolates were designated as FZ06A and FZ16A. Electron microscopy of negatively stained samples revealed the presence of a viral structure with a diameter of 60 to 70 nm in isolated viruses (Supplementary Fig. 1).
The genome sequence of FZ06A and FZ16A strains has been deposited in GenBank under accession No. MF370557 and No. KY761966, respectively. The genomes of FZ06A and FZ16A strains were comprised of 15,319 and 15,320 nucleotides in length. Likewise, FZ06A and FZ16A strains possessed the genetic marker of 1 + 29 AA (1 and 29 non-contiguous AA deletions at positions 481 and 533–561, respectively) deletions in the Nsp2. It is highly similar to a group of highly pathogenic strains of PRRSV previously isolated in China.
Following comparison of the FZ06A and FZ16A strains with 8 other PRRSV isolates deposited in GenBank (Table 3), it was observed that both FZ06A and FZ16A shared the highest nucleotide identity (range, 97.4%–100%) with the JXA1 isolates, 79.8% to 93.7% identity with NADC30- or NADC30-like (NADC30, HNyc15) strains. An 87.4% to 95.4% sequence identity with VR-2332 (North American type), whereas only 55.8% to 71.7% identity with the Lelystad virus (LV, European type), indicating that these two isolate strains belonged to PRRSV type 2. The 5′ UTR and 3′ UTR of FZ06A and FZ16A shared 93.6% to 99.5% and 92.6% to 99.3% nucleotide identity, respectively, with the HP-PRRSVs (BJ-4, HB-1(sh)/2002, and JXA1), and 92.6% to 93.7% and 87.9% to 89.3% nucleotide identities, respectively with the NADC30- or NADC30-like PRRSVs (NADC30 and HNyc15), as well as 62.8% to 97.9% and 71.4% to 96% nucleotide identity, respectively with VR-2332, CH-1a, and LV strains. The ORF1a and ORF1b of FZ06A and FZ16A shared 87.3% to 99.8% identity with the HP-PRRSVs (BJ-4, HB-1(sh)/2002, and JXA1), 79.8% to 88% nucleotide identity with the NADC30 or NADC30-like PRRSV (NADC30 and HNyc15), and 55.8% to 96.4% nucleotide identity with VR-2332, CH-1a, and LV. Whereas, ORF2 to ORF7 were comparatively conserved with identities of 87.4% to 100% with the HP-PRRSVs BJ-4, HB-1(sh)/2002, and JXA1, and 83.3% to 90.9% nucleotide identity with the NADC30- or NADC30-like PRRSVs (NADC30 and HNyc15). In contrast, they only showed 63.3% to 69.5% identify with LV (Table 3).
By comparing the Nsp2 AA sequences of other type 2 PRRSV strains, we found 65.3% to 98% and 65.3% to 92% identities between FZ06A/type 2 PRRSV and FZ16A/type 2 PRRSV, respectively (Table 3). In addition, when compared to other PRRSV strains, the AA mutations in position 307, 361, 373, 482, 486, 514, 677, 712 and 771 were found in Nsp2 of the FZ16A, whereas there was no mutation in Nsp2 of the FZ06A strain.
The AA substitutions R13 and R151 associated with high virulence ability were identified in the predicted GP5 protein of FZ06A strain. The FZ16A strain also shared the AA substitution R13, but it also had a K151 AA substitution. Changes in the AA sequence of potential N-glycosylation sites (NGSs) in GP5 significantly influenced the susceptibility of mutant viruses to virus-neutralizing antibodies. To gain further insight into the genetic evolution of FZ06A and FZ16A, we also analyzed variation of GP5 in potential NGSs by using NetOGlyc1.0 Server. The potential NGSs were detected at six different positions: N30, N33, N34, N35, N44, and N51. The GP5 of the FZ16A strain shared five common NGSs at AA positions 30, 34, 35, 44, and 51; however, the deletion of one NGS at position 33 was detected in any other PRRSV strains. The FZ06A strain shared six common NGSs (Table 4).
In order to determine the phylogenetic relationship between FZ06A and FZ16A strains and other characterized PRRSV isolates, phylogenetic trees were constructed based on the nucleotide sequences of the complete genome, Nsp2, and ORF5 of the FZ06A and FZ16A strains by using MEGA software (ver. 5.1) (Fig. 2). Phylogenetic analysis demonstrated that all PRRSV isolates could be divided into two types. Moreover, the type II isolates could be further divided into four main groups. Group 1 contained four North American strains (VR-2332, RespPRRS MLV, and BJ-4) and group 2 contained some recombination strains (e.g., HNyz15, HENAN-HEB, and CHsx1401). Likewise, group 3 contained several classical Chinese strains (CH-1a and Ch-1R) and group 4 contained some Chinese HP-PRRSV strains (e.g., JXA1, JX143, and SY0608). The phylogenetic trees showed that the FZ16A strain was similar to the Chinese e JX143 strain and the FZ06A strain was similar to the SY0608 strain, which belonged to the group 4.
The rectal temperature and clinical signs of infected animals were recorded daily throughout the experiment, and there were no typical clinical signs in animals from the negative-control group and no remarkable clinical symptoms detected in the inoculated animals until 2 dpi. From 3 dpi onward, almost all inoculated animals presented with depression, low appetite, coughing, sneezing, and diarrhea. The rectal temperature of FZ16A-challenged animals was mildly elevated to 40℃ at 4 or 5 dpi and this persisted for 2 to 5 days; however, rectal temperature did not exceed 40.5℃ throughout the observation period. In comparison with the FZ16A-challenged group, the rectal temperature of FZ06A-challenged animals also had a high temperature. After 4 days, the rectal temperature elevated to 40℃ and persisted at that level for 11 to 12 days, and, in some challenged animals, the temperature exceeded 41℃. The rectal temperature in the FZ06A group was significantly higher than that of the FZ16A and control groups (Fig. 3).
The viral loads in the sera of infected pigs were measured by using an IFA-microtitration infectivity assay. The sera from the FZ06A-infected group showed increased viral titers at 4 dpi, which peaked at 11 dpi. The FZ16A-infected group also showed increased viral titers, and increased viral titers were present for a longer period in the FZ16A-infected than in the FZ06A-infected group (data not shown).
Sera were collected at 1, 3, 7, 10, 14, and 21 dpi to monitor PRRSV-specific antibodies. The effect of antibodies against PRRSV was analyzed using a commercially available IDEXX ELISA. All inoculated pigs became seropositive (S/P ratio > 0.4) at 7 dpi. The S/P ratio then gradually increased until the end of the examination; no PRRSV-specific antibodies were detected in the control animals (Fig. 4).
Virus detection in tissue samples collected from negative controls was negative for the presence of PRRSV, whereas viruses were detected in all evaluated organs of inoculated pigs. All tissue samples from the control and inoculated animals were found to be negative for the presence of PRV, PCV, or CSFV.
All pigs were euthanized and anatomized at 21 dpi. No macroscopic (gross) lesions were observed in the lungs collected from the control animals at necropsy (panel A in Fig. 5). Animals from the FZ06A-infected group exhibited severe multifocal lesions and consolidation in the lungs (panel B in Fig. 5), whereas the FZ16A-infected group only exhibited mild multifocal lesions in the lungs (panel C in Fig. 5).
Under histological examination, none of the lungs from control pigs exhibited any features indicative of acute PRRSV infection (panel A in Fig. 6). However, the lung tissues collected from FZ06A-infected and FZ16A-infected animals displayed interstitial pneumonia with alveolar septal thickening and alveoli disappearance (panels B and C in Fig. 6).
PRRS is one of the highly-ranked serious swine diseases and affects industries related to pigs around the world. In China, PRRS was first reported in 1996. In June 2006, there was an unparalleled outbreak of HP-PRRSV in China. Since then, PRRSV has become widespread and today various types of PRRS have been isolated, and the number of strains is growing rapidly [32]. Traditional control strategies and conventional vaccines have been unable to provide sustainable disease control; therefore, commercial vaccines specifically against the PRRS have been developed. Because of the considerable genetic and antigenic diversities in HP-PRRSV isolates, surveillance and identification of the recently emerged new virus strains have become crucial [29].
In the current study, two PRRSV strains were isolated from a pig farm in Fujian, China, one isolated in 2006 and the other in 2016. The complete nucleotide sequences of the FZ06A and FZ16A strains were determined and then compared with other representative PRRSV strains that had been isolated in China and other countries. Based on genome analysis, the FZ06A and FZ16A strains had a discontinuous deletion of 30 AA (1 AA and 29 AA) in Nsp2, which is suggested to be a marker of a HP-PRRSV [32]. Based on phylogenetic analysis, a high degree of genetic homology was observed among most of the Chinese isolates.
Clinical signs of PRRSV infection developed in pigs challenged with the FZ06A and FZ16A strains. In the FZ16A-infected group, no mortality was observed, but the FZ06A-infected group exhibited high mortality. The absence of mortality detected during the challenge study indicated that the virulence of FZ16A was relatively weak. The low virulence might be related to the evolution of Chinese HP-PRRSV under selective pressure.
The Nsp2 gene exhibited the highest genetic diversity in the genome of PRRSV [1025]. Nsp2 is crucial for viral replication and modulation of host immunity due to its protease activity [535], The HP-PRRSV strains that emerged in 2006 had a unique 30 AA (1 AA + 29AA) deletion in the Nsp2 hyper-variable region. In recent years, a variety of atypical PRRSV strains with new AA deletions or insertions in Nsp2 have been reported [8]. The virulence of these strains differs from their typical PRRSV counterparts. A recent study reported that the 30 AA deletion in the Nsp2-coding region could be considered as a putative marker for high virulence in a low virulence strain [21]. In this study, we detected a discontinuous 30 AA deletion of 1 AA + 29AA in the Nsp2 region of both FZ06A and FZ16A strains at positions 481 and 533 to 561, which is similar to that of Chinese HP-PRRSV isolates such as JXA1, HUN4, and BJ-4. We also observed AA mutations in other positions including 307 (A→V), 361 (G→E), 373 (E→G), 482 (A→V), 486 (L→P), 514 (V→M), 677 (D→N), 712 (K→E), and 771 (I→T) in Nsp2 of FZ16A when compared to HP-PRRSV in China; however, there were no obvious mutations in the Nsp2 of FZ06A when compared to HP-PRRSVs in China. The phylogenetic tree analysis showed that the FZ16A strain had similarity to the Chinese JX143 strain, whereas there were no similar AA mutations in Nsp2 in the FZ16A and JX143 strains. Likewise, these two (JX143 and FZ16A) strains have different levels of virulence. However, further study is essential to determine whether these mutations caused the virulence difference. In a previous study, it was reported that the 30 AA deletion was not associated with the virulence of emerging HP-PRRSVs [39], whereas in our research, both of these two isolate strains had the 30 AA deletion. There were various kinds of mutations in the Nsp2 of FZ16A compared to those in the Nsp2 of FZ06A, and there were obvious differences in experimental inoculation results from the two strains. Thus, we suggest that these mutations may have an influence on the virulence of the FZ16A isolate.
GP5 is a glycosylated viral transmembrane protein, which is responsible for virus attachment to the host cell and contains important immunological domains associated with virus neutralization [20]. It has been reported that GP5 is the least conserved ORF of PRRSV, and AA sequence variations in GP5 are mainly observed in the extra virion domain of the protein, which has been shown to be responsible for cell attachment. N-glycosylation of GP5 might be critical for proper functioning of the protein. Potential involvement of NGSs in GP5 has been observed in viral immune evasion and virus-neutralizing antibody responses [339]. Glycosylate H38 (L/F) 39 residues in this domain are considered to be critical to the immunogenicity of this epitope [27]. Both major and minor viral envelope glycoproteins can help PRRSV to escape, block, or minimize the viral neutralizing antibody response [34].
Glycoprotein 5 is essential for virus infectivity due to its significant genetically variable effect on structural proteins of PRRSV. Residues 13 and 151 in the GP5 proteins of PRRSVs have been shown to be associated with virulence [1]. Compared to the first Chinese strain, CH-1a, the other PRRSV strains exhibit variation in their potential glycosylation level or sites. Based on our results, residues 13 and 151 in the GP5 proteins of PRRSVs are associated with virulence. With the changing and increasing number of NGSs, a low value of antigenicity was obtained. In our study, potential NGSs were also observed in the GP5 of FZ06A and FZ16A strains at AA positions 30, 34, 35, 44, and 51. Interestingly, the FZ06A shared six common NGSs with other HP-PRRSVs in China (Table 4); regardless, there was one possible NGS in the FZ16A strain at position 33 (N33→K33) that, thus far, has not been observed in any other PRRSV strains. Whether this mutation is associated with virulence remains to be determined.
To confirm the pathogenicity of the FZ06A and FZ16A strains, SPF pigs were inoculated with 105 TCID50 of FZ06A or FZ16A. At 3 to 11 dpi, all inoculated animals presented clinical signs of PRRSV infection. In the FZ16A-infected group, there was no mortality, while in FZ06A-infected group four of five inoculated pigs died between 7 and 21 dpi. Development of specific antibodies in inoculated pigs was detected at 7 dpi by ELISA, and PCR analysis showed PRRSV-positive tissue samples including the lung, spleen, kidney, liver, and lymph nodes. Pigs in the control group remained seronegative throughout the experiment. Histological examination revealed that the lungs from the FZ16A-infected animals developed interstitial pneumonia with slighter alveolar septal thickening, alveoli disappearance, and atrophy than the FZ06A-infected animals. It has been reported that the age of piglets might be associated with their resistance against PRRSV infection [19]. The FZ06A and FZ16A strains produced different mortality; thus, we conclude that after ten years evolution of Chinese HP-PRRSV under selective pressure, the FZ16A strain has adapted to the environment to a greater extent than FZ06A.
In summary, the two PRRSV isolate strains (FZ06A and FZ16A) isolated in 2006 and 2016, respectively, showed similar genomic characteristics with HP-PRRSV in China but different mortality effects; the virulence difference indicating that HPPRRSV strains prevailing in China may have gradually adapted during repeated passages in animals. Furthermore, the types of mutation in Nsp2 and the unique mutation in GP5 are crucial to the virulence of HP-PRRSVs. The high degree of genetic and antigenic diversity among field isolates underline the complexity to be overcome when attempting to control and eradicate PRRS in China. This study provides useful information on the evolutionary characteristics of Chinese PRRSVs; such information could be utilized as reference material in the development of future live attenuated vaccines against this economically devastating disease.
Figures and Tables
Fig. 1
Cytopathic effects of isolated porcine reproductive and respiratory syndrome virus cultured in Marc-145 cells. Infected Marc-145 cells became round and aggregated into clusters. (A) Non-infected Marc-145. (B) Virus-infected Marc-145 cells. Scale bars = 200 µm (A and B).

Fig. 2
Phylogenetic tree of 24 porcine reproductive and respiratory syndrome virus (PRRSV) isolates based on analysis of nucleotide sequences of the complete genomic sequences (A), Nsp2 (B), and ORF5 (C) of PRRSV strains by applying the distance-based neighbor-joining method in MEGA software (ver. 5.1) [31]. Bootstrap values were calculated on 1,000 replicates of the alignment.

Fig. 3
Mean rectal temperature in negative-control pigs and pigs infected experimentally with FZ06A and FZ16A porcine reproductive and respiratory syndrome virus. Significant differences between FZ06A-infected, FZ16A-infected and negative-control pigs were observed at most of the time point.

Fig. 4
Serum antibody levels over time. Values represent average titers from five animals. Dotted line at 0.4 sampleto-positive (S/P) ratio designates threshold value above which titers were considered positive for anti-porcine reproductive and respiratory syndrome virus antibodies. Data from groups of five pigs assessed by using one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001).

Fig. 5
Macroscopic observation of the lungs of the control (A) and highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV)-infected (B and C) pigs. No macroscopic (gross) lesions were recorded in the lungs collected from the control animals. (B) Severe multifocal lung lesions and consolidation of lung tissue in the HP-PRRSV FZ06A-infected group. (C) Mild multifocal lung lesions and consolidation of lung tissue in the HP-PRRSV FZ16A-infected group.

Fig. 6
Histopathological lesions in the lung of control (A) and highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infected (B and C) pigs. Histological lesions were examined in the HP-PRRSV FZ06A- and FZ16A-infected groups and the challenge control by using H&E staining. (A) All lungs from control pigs failed to exhibit features of acute PRRSV infection. (B) Lung tissues collected from the HP-PRRSV FZ06A-infected pigs developed severe interstitial pneumonia with alveolar septal thickening and alveoli disappearance. (C) The lung tissues collected from HP-PRRSV FZ16A-infected pigs developed mild interstitial pneumonia and alveoli disappearance. Scale bars = 200 µm (A–C).
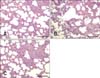
Table 1
Primers used for amplification and sequencing of gene fragments of PRRSV strains FZ06A and FZ16A

Acknowledgments
This work was supported by the Agricultural Science and Technology Innovation Program (No. ASTIP-IAS-11) and the Fund on Basic Scientific Research Project of Nonprofit Central Research Institutions, Institute of Animal Sciences in CAAS (2014ywf-zd-1). We thank the University of Liège-Gembloux Agro-Bio Tech and more specifically the research platform Agriculture Is Life for funding the scientific stay in Belgium that made this paper possible.
References
1. Allende R, Kutish GF, Laegreid W, Lu Z, Lewis TL, Rock DL, Friesen J, Galeota JA, Doster AR, Osorio FA. Mutations in the genome of porcine reproductive and respiratory syndrome virus responsible for the attenuation phenotype. Arch Virol. 2000; 145:1149–1161.

2. An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg Infect Dis. 2011; 17:1782–1784.

3. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.

4. Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 1992; 4:127–133.

5. Chen Z, Zhou X, Lunney JK, Lawson S, Sun Z, Brown E, Christopher-Hennings J, Knudsen D, Nelson E, Fang Y. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol. 2010; 91:1047–1057.

6. Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992; 4:117–126.

7. Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000; 145:659–688.

8. Du Y, Lu Y, Qi J, Wu J, Wang G, Wang J. Complete genome sequence of a moderately pathogenic porcine reproductive and respiratory syndrome virus variant strain. J Virol. 2012; 86:13883–13884.

9. Fan B, Wang H, Bai J, Zhang L, Jiang P. A novel isolate with deletion in GP3 gene of porcine reproductive and respiratory syndrome virus from mid-eastern China. Biomed Res Int. 2014; 2014:306130.
10. Fang Y, Fang L, Wang Y, Lei Y, Luo R, Wang D, Chen H, Xiao S. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-κB activation. Virol J. 2012; 9:83.

11. Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RR. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004; 100:229–235.

12. Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol. 2011; 92:1097–1106.

13. Forsberg R, Storgaard T, Nielsen HS, Oleksiewicz MB, Cordioli P, Sala G, Hein J, Bøtner A. The genetic diversity of European type PRRSV is similar to that of the North American type but is geographically skewed within Europe. Virology. 2002; 299:38–47.

14. Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995; 32:648–660.

15. Huang C, Zhang Q, Feng WH. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015; 202:101–111.

16. Jiang P, Chen PY, Dong YY, Cai JL, Cai BX, Jiang ZH. Isolation and genome characterization of porcine reproductive and respiratory syndrome virus in P.R. China. J Vet Diagn Invest. 2000; 12:156–158.

17. Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol. 2011; 92:1107–1116.

18. Karniychuk UU, Geldhof M, Vanhee M, Van Doorsselaere J, Saveleva TA, Nauwynck HJ. Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res. 2010; 6:30.

19. Ladinig A, Detmer SE, Clarke K, Ashley C, Rowland RR, Lunney JK, Harding JC. Pathogenicity of three type 2 porcine reproductive and respiratory syndrome virus strains in experimentally inoculated pregnant gilts. Virus Res. 2015; 203:24–35.

20. Li B, Xiao S, Wang Y, Xu S, Jiang Y, Chen H, Fang L. Immunogenicity of the highly pathogenic porcine reproductive and respiratory syndrome virus GP5 protein encoded by a synthetic ORF5 gene. Vaccine. 2009; 27:1957–1963.

21. Li Y, Xue C, Wang L, Chen X, Chen F, Cao Y. Genomic analysis of two Chinese strains of porcine reproductive and respiratory syndrome viruses with different virulence. Virus Genes. 2010; 40:374–381.

22. Ma H, Kien F, Manière M, Zhang Y, Lagarde N, Tse KS, Poon LL, Nal B. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J Virol. 2012; 86:1789–1801.

24. Murtaugh MP, Faaberg KS, Laber J, Elam M, Kapur V. Genetic variation in the PRRS virus. Adv Exp Med Biol. 1998; 440:787–794.

25. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999; 73:270–280.

26. Ni J, Yang S, Bounlom D, Yu X, Zhou Z, Song J, Khamphouth V, Vatthana T, Tian K. Emergence and pathogenicity of highly pathogenic Porcine reproductive and respiratory syndrome virus in Vientiane, Lao People's Democratic Republic. J Vet Diagn Invest. 2012; 24:349–354.

27. Ostrowski M, Galeota JA, Jar AM, Platt KB, Osorio FA, Lopez OJ. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol. 2002; 76:4241–4250.

29. Shi M, Lam TT, Hon CC, Murtaugh MP, Davies PR, Hui RK, Li J, Wong LT, Yip CW, Jiang JW, Leung FC. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010; 84:8700–8711.

30. Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol. 1998; 79:961–979.

31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.

32. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007; 2:e526.

33. Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. 2007; 13:1434–1436.

34. Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol. 2011; 85:5555–5564.

35. Wang FX, Song N, Chen LZ, Cheng SP, Wu H, Wen YJ. Non-structural protein 2 of the porcine reproductive and respiratory syndrome (PRRS) virus: a crucial protein in viral pathogenesis, immunity and diagnosis. Res Vet Sci. 2013; 95:1–7.

36. Wang X, Marthaler D, Rovira A, Rossow S, Murtaugh MP. Emergence of a virulent porcine reproductive and respiratory syndrome virus in vaccinated herds in the United States. Virus Res. 2015; 210:34–41.

37. Zhang G, Lu W, Chen Y, Zhu L, Wei Z, Li Z, Sun B, Xie Q, Bi Y, Ma J. Complete genome sequence of two variant porcine reproductive and respiratory syndrome viruses isolated from vaccinated piglets. J Virol. 2012; 86:11396–11397.

38. Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009; 145:97–105.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download