Abstract
Background
Following discontinuation of the recombinant immunoblot assay (RIBA), the only available supplementary test for the detection of hepatitis C virus (HCV) is the nucleic acid amplification test (NAAT). However, the NAAT does not adequately detect past HCV. Consequently, it is hard to distinguish between past HCV infection and biological false positivity with an anti-HCV result alone. We assessed the diagnostic performance of two immunoassays: the ARCHITECT anti-HCV chemiluminescent microparticle immunoassay (CMIA; Abbott Diagnostics, Wiesbaden, Germany) and the Access HCV Ab PLUS chemiluminescent immunoassay (CIA; Bio-Rad, Marnes-la-Coquette, France). We also explored an optimized algorithm to determine the anti-HCV results.
Methods
We tested 126,919 patients and 44,556 individuals who underwent a medical checkup. RIBA and NAAT were conducted for samples that tested anti-HCV-positive using CMIA and CIA. We assessed the optimal signal-to-cutoff (S/CO) ratio in HCV-positive samples.
Results
In total, 1,035 blood samples tested anti-HCV-positive. Of these, RIBA was positive in 512, indeterminate in 160, and negative in 363 samples. One hundred sixty-five samples were NAAT-positive. Diagnostic sensitivity and positive predictive value (PPV) were 96.7% and 52.1%, respectively, for CMIA, and 94.7% and 72.3%, respectively, for CIA. The optimal S/CO ratio was 5.2 for CMIA and 2.6 for CIA at 95% PPV. In total, 286 samples tested positive in CMIA and 444 in CIA, while 443 samples tested positive in both assays.
Anti-HCV is detectable 8–12 weeks after hepatitis C virus (HCV) infection [1]. As antibodies against HCV are not neutralizing, they are detected not only in chronic hepatitis C patients but also in the majority of recovered patients. Thus, anti-HCV positivity cannot clearly distinguish current, active infection from past infection [2], and positive anti-HCV results can indicate active HCV infection (acute or chronic), past, resolved HCV infection, or a false-positive anti-HCV result [34]. False positives may occur under conditions of myeloma, rheumatoid arthritis, liver cirrhosis, cancer, or autoimmune diseases (collagenous, autoimmune hepatitis) [56]. Anti-HCV testing by third-generation anti-HCV ELISA shows a high false-positive rate of 15–60% (mean 35%) in low-risk groups for HCV infection, such as blood donors and healthcare workers as well as the general population [7891011121314]. To overcome this issue, efforts have been made to reduce false-positive results by adjusting the signal-to-cutoff (S/CO) ratio [615161718] or by conducting additional tests, such as the nucleic acid amplification test (NAAT) or the recombinant immunoblot assay (RIBA) [192021]. In 2003, the US Centers for Disease Control and Prevention recommended a S/CO ratio for anti-HCV screening tests and a diagnostic algorithm that involves NAAT and RIBA [19]. In July 2013, an updated HCV testing algorithm involving sequential testing of anti-HCV screening-positive samples with HCV RNA using only the NAAT was published [20]. The most important change to the testing algorithm was the withdrawal of the US Food and Drug Administration approval of RIBA, a supplemental test for anti-HCV. Therefore, it became challenging to discriminate false-positive anti-HCV results from past, resolved HCV infection by the NAAT alone.
Past, resolved HCV infection and false-positive anti-HCV results are problematic in two aspects. First, when a patient tests positive for anti-HCV, especially during a medical checkup, clinicians will strive to determine false positivity not only through laboratory approaches but also by using imaging techniques, such as ultrasonography and computed tomography. As HCV requires continuous monitoring since reinfection can occur even after recovery [22], false positivity may have significant consequences for the patient in terms of time and cost. Second, patients with a false-positive anti-HCV result are incorrectly banned from donating blood. However, this is not an issue for past, resolved HCV infection patients. In retrospective studies [2324], there were no confirmed cases of infection from transfusion of blood that tested positive for anti-HCV, negative or indeterminate for RIBA, that is, false-positive anti-HCV.
Thus, accurate interpretation of anti-HCV-positive results and a clear distinction between past, resolved HCV infection and a false-positive result is needed. We performed both the NAAT and RIBA as confirmatory tests on samples that showed positive results of anti-HCV by two immunoassay (IA) systems. On the basis of the test results, we assessed the diagnostic performance of each IA based on the S/CO ratio, and explored an algorithm with optimal efficiency that assesses the anti-HCV results of one or two IAs.
This retrospective study was performed between July 2013 and June 2016 at Seoul National University Bundang Hospital, Seongnam, Korea. We tested 126,919 patients and 44,556 individuals from those undergoing a medical checkup for anti-HCV using the ARCHITECT (Abbott Diagnostics, Wiesbaden, Germany) and Access (Bio-Rad, Marnes-la-Coquette, France) systems. All positive anti-HCV serum samples were tested on each assay repeatedly, and the NAAT and RIBA were performed as supplemental tests to confirm positive anti-HCV results. As of April 2014, the results were reported as positive when a S/CO ratio ≥5 in ARCHITECT and ≥3 in Access were achieved simultaneously, without any additional supplemental tests.
In total, 2,388 (1.39%) samples tested positive in the anti-HCV screening. We evaluated the 1,035 samples that remained after applying the following exclusion criteria: patients in the follow-up phase, no NAAT or RIBA results owing to lack of sample, and results with S/CO ratio of ≥5 in ARCHITECT and ≥3 in Access after April 2014. This study was approved by the Seoul National University Bundang Hospital institutional review board (B-1609/361-103). The need for written informed consent was waived.
For patient testing, we used the ARCHITECT anti-HCV chemiluminescent microparticle IA (CMIA) on an ARCHITECT i2000 analyzer. For testing of the medical checkup population, we used the Access HCV Ab PLUS chemiluminescent IA (CIA) on an Access 2 analyzer. The ARCHITECT CMIA employs recombinant antigens to c100-3 (NS4) (yeast-derived), a fusion protein of two noncontiguous coding regions of NS3, and a core produced in Escherichia coli and referred to as HCr43 (HCV Chiron 43) for antibody capture. The Access HCV Ab PLUS CIA includes paramagnetic particles coated with recombinant proteins (NS3/NS4) and capsid peptide suspended in Tris buffer. The S/CO ratio was recorded directly from the automated system. As per the manufacturer's guidelines, a test result was considered positive when the S/CO ratio was ≥1.
The NAAT for HCV RNA was performed using the Abbott RealTime HCV PCR (Abbott Diagnostics, Illinois, USA). The limit of detection of the RealTime HCV PCR assay was 30 IU/mL, with 0.2 mL used for sample preparation. RIBA was conducted using the HCV BLOT 3.0 Strip Immunoblot Assay (MP Diagnostics, Illkirch Cedex, France). The nitrocellulose strips contain four recombinant HCV proteins from both structural (core) and nonstructural (NS3, NS4, and NS5) viral antigen regions. For this assay, the intensity of the colored bands on a nitrocellulose strip is proportional to the amount of bound antibody and was graded as negative, ±, 1+, 2+, 3+, or 4+, according to the manufacturer's instructions. The positive results are either >1+ reactivity with at least 2 HCV antigens or 2+ reactivity to the core band only. An indeterminate result was defined as any single HCV band of 1+ or greater reactivity that did not meet the positive criteria. A negative result was defined as no bands of ≥1+. The tests were carried out according to the manufacturer's instructions.
Samples that tested positive for anti-HCV and for the NAAT were considered true positive, with present HCV infection. Negative NAAT with positive RIBA result indicated true positive and past, resolved HCV infection. Other results (negative or indeterminate RIBA results) were regarded as biological false-positive anti-HCV results [19].
We used the Wilcoxon signed rank test in SPSS ver. 22.0 (IBM Co., Armonk, NY, USA) to compare the results of a repeat test and of the IA. P values of <0.05 from two-sided tests were considered statistically significant. Sensitivity and positive predictive value (PPV) were presented as percentages. We used the Wilson score method [25] without continuity correction in Microsoft Excel 2010 software (Microsoft Corporation, Redmond, WA, USA) to calculate a 95% confidence interval (CI) for a proportion. We calculated the mean and standard deviation (SD) of anti-HCV S/CO values according to the NAAT and RIBA results.
Among the 1,035 samples with positive anti-HCV, RIBA was positive in 512 (47.9%; 163 were NAAT positive), indeterminate in 160 (15.2%; none were NAAT positive), and negative in 363 (36.9%; two were NAAT positive) samples. The results of the two anti-HCV assays were discordant in 448 samples (41.9%); among these discordant samples, CMIA-positive and CIA-negative results were observed in 84 samples, and CIA-positive and CMIA-negative results were observed in 364 samples.
We determined the optimal values for the S/CO ratios of CMIA and CIA results that simultaneously satisfied PPV ≥95% and false-positive rate <5% to be 5.2 and 2.6, respectively (Tables 1 and 2; Fig. 1).
On the basis of these results, we established an algorithm for screening anti-HCV with two different IAs and categorized the results in groups A to F by the optimal S/CO ratio for each IA (Fig. 2). In total, 267 samples met the criteria for the S/CO ratio concurrently in each IA, with a PPV of 99.6% (1 false-positive anti-HCV), whereas 443 samples met only one criterion for the S/CO ratio, with a PPV of 94.8% (23 false-positive anti-HCV). Sequencing before and after the two IAs did not affect the results of groups A, B, and E. However, depending on the performance of each IA, a different number of samples should have been tested with a supplemental test (group F).
We investigated two models for detecting anti-HCV positivity. The first model uses only one IA for anti-HCV testing. In this case, the results are reported simply in reference to the S/CO with 95% PPV: a higher value is reported as positive and a lower value as weak positive. Weak positives should be tested for false positivity through an additional supplemental test. S/CO values with 95% PPV were 5.2 for CMIA and 2.6 for CIA. Thus, the CMIA has a higher false-positive rate and a two-fold greater S/CO value than CIA. Previous studies have also reported high false-positive rates for anti-HCV tests using the ARCHITECT system [152627]. The second model uses two different IAs for anti-HCV testing, which allows for mutual complementation with a cross-check comparison between test results. If the first IA shows a positive anti-HCV result, then the second IA may be used to re-examine and report the results by each S/CO ratio.
The two models delineated above did not use a repeat test or a supplemental test such as HCV NAAT. Most manufacturers recommend retesting with a duplicate if the anti-HCV test shows a positive result. Some studies report cases of a repeat test contributing to the final evaluation [16]. However, in the present study, of the 1,035 samples that were reflex-tested before applying the exclusion rule in CMIA, nine cases of discrepancy were identified under the S/CO 1 criterion, all of which were negative for HCV NAAT and RIBA. Therefore, repeat tests did not affect the results. In case of CIA, a total of 172 samples were repeatedly tested, but none of the samples showed different results. Although the repeat test may be used to identify errors resulting from the mix-up of samples as well as sampling and handling errors [28], our study showed that repeat tests are not necessary. This is because the biological false reaction still showed a result within the range of tolerable bias (data not shown), even after a repeat test with the same IA. In previous studies, a higher anti-HCV S/CO ratio indicated current infection and positive NAAT results [2129]. This study also showed that the NAAT-positive samples had average S/CO values >2-fold higher than those of negative samples (Table 3 and Fig. 3).
To determine HCV infection in the case of low S/CO ratio, for which determination is difficult, we reviewed the medical records of 43 patients with indeterminate RIBA results. There were eight (18.6%) true HCV-infected patients (resolved), and the rest were eight cancer patients, nine medical checkup patients, and 18 patients with other illnesses. S/CO ratios <2 were observed in 30 (70%) patients, and no patients were positive for the NAAT, which is in accordance with a previous report [5]. For patients with past, resolved HCV infection, the S/CO ratio declined from >10 to 1–2 over time. One patient showed a marked decline in the S/CO ratio to <2 within only two years. A high S/CO ratio with a negative NAAT may be regarded as a resolved infection [30]. However, in the patients with spontaneous recovery after infection without subjective symptoms (15–45%) [4], it is difficult to distinguish false-positive results from low S/CO ratio solely on the basis of their medical history, as they overlap with the false-positive results at low S/CO ratios. Therefore, supplemental tests can assist the clinician in decision-making, which may be crucial. However, since the only currently available laboratory test is the NAAT, which is not highly indicative, S/CO ratio analysis is the most important component. The anti-HCV S/CO ratio and HCV infection are related, with a higher S/CO ratio representing true infection. However, no correlation could be ascertained between a true HCV infection and low S/CO ratio. Furthermore, anti-HCV results have many false-positive factors. It is difficult to differentiate false-positive from positive anti-HCV results using the S/CO ratio alone. Some biochemical tests (e.g., alanine aminotransferase, albumin, cholesterol) are helpful in diagnosing HCV infection but are only distinctive in acute HCV infection (data not shown). In some studies, however, there was no difficulty in diagnosing HCV infection despite the permanent discontinuation of RIBA [30]. Our study demonstrated that positive results would not seem to be conclusive of HCV infection without supplemental RIBA testing.
A limitation of our study was the potential selection bias. Our study population was divided into two groups. One group consisted of hospital patients who were tested by CMIA, and the other group comprised subjects who underwent medical checkup by CIA. Because the performances of the two IA are different, the detection rate of anti-HCV-positive samples may be different in the two groups. However, we tried to analyze as many anti-HCV-positive samples as possible, and the detected anti-HCV-positive samples were independently analyzed by CMIA and CIA.
In conclusion, it is not yet possible to conclusively determine HCV infection (active or resolved) through NAAT or by adjusting the S/CO ratio. Furthermore, it is challenging to propose a standard S/CO ratio, because the performance of each IA in terms of S/CO ratio varies across manufacturers [151718,
21]. However, this study is significant because it sought to elucidate the role of the S/CO ratio by referring to the NAAT, RIBA, and medical records to determine as many true positives as possible from anti-HCV-positive results using one or two IAs.
References
1. Ghany MG, Strader DB, Thomas DL, Seeff LB. American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009; 49:1335–1374. PMID: 19330875.
2. Korean Association for the Study of the Liver. Chronic Hepatitis C Clinical Practice Guideline [guidelines]. Updated on Apr, 2018. http://www.kasl.org/bbs/index.html?code=guide&category.
3. Pawlotsky JM. Use and interpretation of virological tests for hepatitis C. Hepatology. 2002; 36(Suppl 1):S65–S73. PMID: 12407578.
4. World Health Organization. Guidelines for the Screening Care and Treatment of Persons with Chronic Hepatitis C Infection: Updated Version [guidelines]. Updated on Apr, 2018. http://www.who.int/hepatitis/publications/hepatitis-c-guidelines-2016/en/.
5. Pereira FM, Zarife MA, Reis EA, G Reis M. Indeterminate RIBA results were associated with the absence of hepatitis C virus RNA (HCV-RNA) in blood donors. Rev Soc Bras Med Trop. 2014; 47:12–17. PMID: 24603731.
6. Berger A, Rabenau H, Allwinn R, Doerr HW. Evaluation of the new ARCHITECT anti-HCV screening test under routine laboratory conditions. J Clin Virol. 2008; 43:158–161. PMID: 18635393.
7. Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999; 341:556–562. PMID: 10451460.
8. Conry-Cantilena C, VanRaden M, Gibble J, Melpolder J, Shakil AO, Viladomiu L, et al. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996; 334:1691–1696. PMID: 8637513.
9. Goetz AM, Ndimbie OK, Wagener MM, Muder RR. Prevalence of hepatitis C infection in health care workers affiliated with a liver transplant center. Transplantation. 1995; 59:990–994. PMID: 7535961.
10. Gunn RA, Murray PJ, Ackers ML, Hardison WG, Margolis HS. Screening for chronic hepatitis B and C virus infections in an urban sexually transmitted disease clinic: rationale for integrating services. Sex Transm Dis. 2001; 28:166–170. PMID: 11289199.
11. Hyams KC, Riddle J, Rubertone M, Trump D, Alter MJ, Cruess DF, et al. Prevalence and incidence of hepatitis C virus infection in the US military: a seroepidemiologic survey of 21,000 troops. Am J Epidemiol. 2001; 153:764–770. PMID: 11296148.
12. Kleinman S, Alter H, Busch M, Holland P, Tegtmeier G, Nelles M, et al. Increased detection of hepatitis C virus (HCV)-infected blood donors by a multiple-antigen HCV enzyme immunoassay. Transfusion. 1992; 32:805–813. PMID: 1471243.
13. Montecalvo MA, Lee MS, DePalma H, Wynn PS, Lowenfels AB, Jorde U, et al. Seroprevalence of human immunodeficiency virus-1, hepatitis B virus, and hepatitis C virus in patients having major surgery. Infect Control Hosp Epidemiol. 1995; 16:627–632. PMID: 8601681.
14. Roome AJ, Hadler JL, Thomas AL, Migicovsky B, Roth R, Boraz M, et al. Hepatitis C virus infection among firefighters, emergency medical technicians, and paramedics--selected locations, United States, 1991-2000. MMWR Morb Mortal Wkly Rep. 2000; 49:660–665. PMID: 10943253.
15. Maasoumy B, Bremer B, Raupach R, Lehmann P, Manns MP, Cornberg M, et al. How to interpret borderline HCV antibody test results: a comparative study investigating four different anti-HCV assays. Viral Immunol. 2014; 27:7–13. PMID: 24494968.
16. Acar A, Kemahli S, Altunay H, Kosan E, Oncul O, Gorenek L, et al. The significance of repeat testing in Turkish blood donors screened with HBV, HCV and HIV immunoassays and the importance of S/CO ratios in the interpretation of HCV/HIV screening test results and as a determinant for further confirmatory testing. Transfus Med. 2010; 20:152–159. PMID: 20059750.
17. Moretti M, Pieretti B, Masucci A, Sisti D, Rocchi M, Delprete E. Role of signal-to-cutoff ratios in hepatitis C virus antibody detection. Clin Vaccine Immunol. 2012; 19:1329–1331. PMID: 22695164.
18. Stramer SL, Dodd RY, Brodsky JP. The value of screening signal-to-cutoff ratios for hepatitis C virus antibody confirmation. Transfusion. 2013; 53:1497–1500. PMID: 23176156.
19. Alter MJ, Kuhnert WL, Finelli L. Centers for Disease Control and Prevention. Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003; 52:1–13. 15quiz CE1-4.
20. Centers for Disease Control and Prevention. Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013; 62:362–365. PMID: 23657112.
21. Lai KK, Jin M, Yuan S, Larson MF, Dominitz JA, Bankson DD. Improved reflexive testing algorithm for hepatitis C infection using signal-to-cutoff ratios of a hepatitis C virus antibody assay. Clin Chem. 2011; 57:1050–1056. PMID: 21566071.
22. AASLD/IDSA HCV Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015; 62:932–954. PMID: 26111063.
23. Vrielink H, Reesink HW, Zaaijer HL, Scholten E, Kremer LC, Cuypers HT, et al. Look-back of anti-HCV ELISA-positive, HCV-RNA PCR-negative donors and recipients of their blood products. Vox Sang. 1997; 72:67–70. PMID: 9088071.
24. Vrielink H, van der Poel CL, Reesink HW, Zaaijer HL, Scholten E, Kremer LC, et al. Look-back study of infectivity of anti-HCV ELISA-positive blood components. Lancet. 1995; 345:95–96. PMID: 7815889.
25. Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998; 17:857–872. PMID: 9595616.
26. Ballani N, Gupta E. Low signal-to-cutoff ratio (S/Co) in the diagnosis of hepatitis C: a diagnostic dilemma? Indian J Gastroenterol. 2015; 34:413–414. PMID: 26498021.
27. Sommese L, Iannone C, Cacciatore F, De Iorio G, Napoli C. Comparison between screening and confirmatory serological assays in blood donors in a region of South Italy. J Clin Lab Anal. 2014; 28:198–203. PMID: 24478048.
28. Pawlotsky JM, Lonjon I, Hezode C, Raynard B, Darthuy F, Remire J, et al. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998; 27:1700–1702. PMID: 9620345.
29. Contreras AM, Ochoa-Jimenez RJ, Celis A, Mendez C, Olivares L, Rebolledo CE, et al. High antibody level: an accurate serologic marker of viremia in asymptomatic people with hepatitis C infection. Transfusion. 2010; 50:1335–1343. PMID: 20088833.
30. Gong S, Schmotzer CL, Zhou L. Evaluation of quantitative real-time PCR as a hepatitis C virus supplementary test after RIBA discontinuation. J Clin Lab Anal. 2016; 30:418–423. PMID: 26499369.
Fig. 1
Results of the RIBA and NAAT with optimal S/CO associated with PPV of 95%: (A) CMIA, (B) CIA.
Abbreviations: RIBA, recombinant immunoblot assay; P, positive; I, indeterminate; N, negative; NAAT, nucleic acid amplification test; S/CO, signal to cut-off ratio; PPV, positive predictive value; CMIA, chemiluminescent microparticle immunoassay; CIA, chemiluminescent immunoassay.
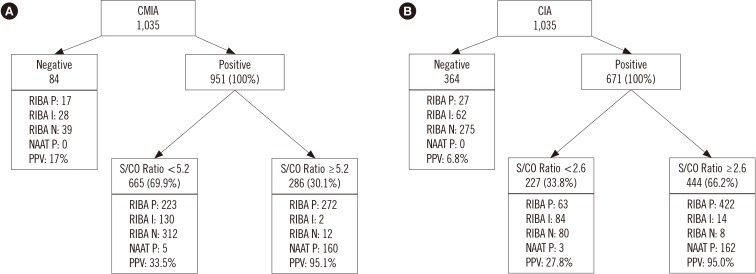
Fig. 2
Algorithm for screening anti-HCV with CMIA and CIA; groups A to F categorized by optimal values of each S/CO ratio: (A) CMIA to CIA, (B) CIA to CMIA.
*Group B does not include Group A.
Abbreviations: RIBA, recombinant immunoblot assay; P, positive; I, indeterminate; N, negative; NAAT, nucleic acid amplification test; S/CO, signal to cut-off ratio; PPV, positive predictive value; HCV, Hepatitis C virus; CMIA, chemiluminescent microparticle immunoassay; CIA, chemiluminescent immunoassay.
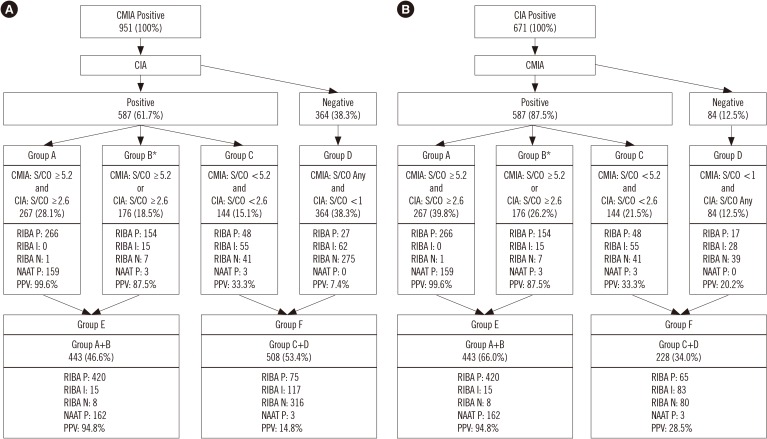
Fig. 3
Distribution of S/CO ratio according to the HCV infection states: (A) CMIA, (B) CIA. Bars represent the interquartile range, and the line within box represent the median. The I bars represent the 2.5th and 97.5th percentiles. Data are presented as mean standard deviation.
Abbreviations: HCV, Hepatitis C virus; S/CO, signal to cut-off ratio; CMIA, chemiluminescent microparticle immunoassay; CIA, chemiluminescent immunoassay.
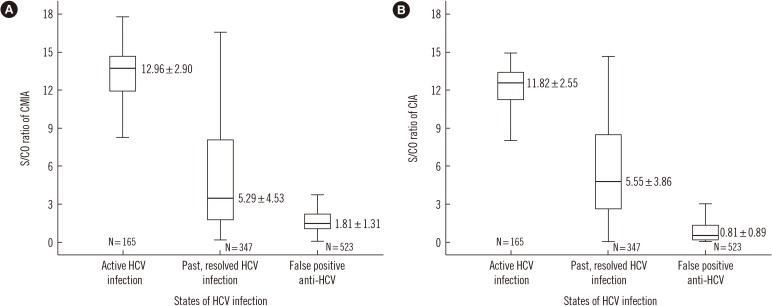
Table 1
Results of RIBA, NAAT, diagnostic performance, and their relationship with the different cutoff points by the S/CO ratios of CMIA
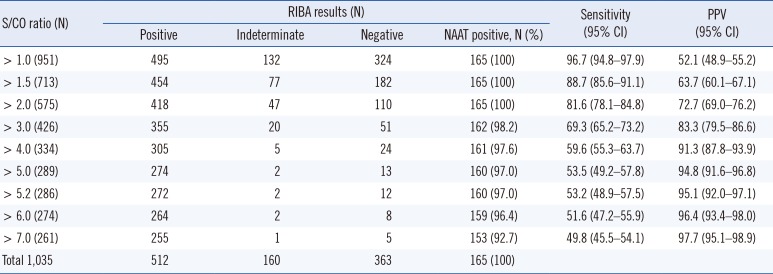
Table 2
Results of RIBA, NAAT, diagnostic performance, and their relationship with the different cutoff points by the S/CO ratios of CIA
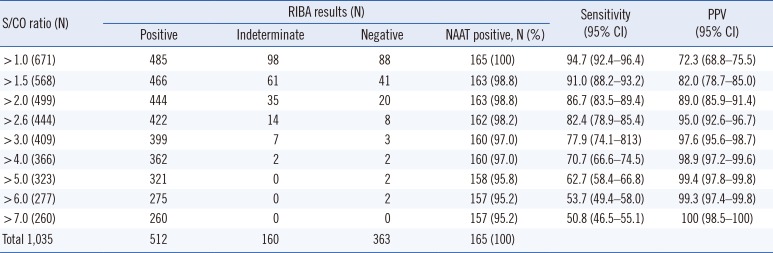
Table 3
S/CO ratios of anti-HCV results by NAAT and RIBA
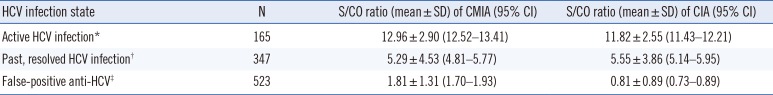
*NAAT positive; †NAAT negative, RIBA positive, ‡NAAT negative, RIBA indeterminate or negative
Abbreviations: CMIA, chemiluminescent microparticle immunoassay; CIA, chemiluminescent immunoassay; RIBA, recombinant immunoblot assay; NAAT, nucleic acid amplification test; S/CO, signal to cut-off; HCV, hepatitis C virus.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download