Abstract
Metabolic reprogramming of cancer cell is one of essential hallmarks of cancer. Otto Warburg first demonstrated that cancer cells utilized more glucose and enhanced glycolytic pathway in the presence of oxygen in 1926. Scientific observations of basic and clinical research in several decades supported that cancer-specific metabolism can be an emerging target for treatment of cancer. Metabolic reprogramming is regulated by both oncogenic signaling and tumor suppressor genes associated with critical signaling pathways in metabolism. These changes provided energy, substrates for cell growth and proliferation, favoring microenvironment, and important for redox balancing for cancer cells. Recent advance of several tools for evaluation comprehensive metabolic profiles of cancer cells provided us to identification of metabolic Achilles' heel of cancers including thyroid cancer. This approach can be a useful strategy for advance in treatment of cancer patients.
Go to : 
References
1. American Thyroid Association (ATA) Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid Cancer. Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009; 19(11):1167–214.

2. Shoup M, Stojadinovic A, Nissan A, Ghossein RA, Freedman S, Brennan MF, et al. Prognostic indicators of outcomes in patients with distant metastases from differentiated thyroid carcinoma. J Am Coll Surg. 2003; 197(2):191–7.

3. Schlumberger M, Brose M, Elisei R, Leboulleux S, Luster M, Pitoia F, et al. Definition and management of radioactive iodine-refractory differentiated thyroid cancer. Lancet Diabetes Endocrinol. 2014; 2(5):356–8.

4. Durante C, Haddy N, Baudin E, Leboulleux S, Hartl D, Travagli JP, et al. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: benefits and limits of radioiodine therapy. J Clin Endocrinol Metab. 2006; 91(8):2892–9.

5. Brose MS, Nutting CM, Jarzab B, Elisei R, Siena S, Bastholt L, et al. Sorafenib in radioactive iodine-refractory, locally advanced or metastatic differentiated thyroid cancer: a randomised, double-blind, phase 3 trial. Lancet. 2014; 384(9940):319–28.

6. Schlumberger M, Tahara M, Wirth LJ, Robinson B, Brose MS, Elisei R, et al. Lenvatinib versus placebo in radioiodinerefractory thyroid cancer. N Engl J Med. 2015; 372(7):621–30.

7. Fojo T, Parkinson DR. Biologically targeted cancer therapy and marginal benefits: are we making too much of too little or are we achieving too little by giving too much? Clin Cancer Res. 2010; 16(24):5972–80.

9. Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012; 12(7):487–93.

10. Stincone A, Prigione A, Cramer T, Wamelink MM, Campbell K, Cheung E, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2014. [Epub ahead of print].

11. Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011; 10(9):671–84.

12. Sborov DW, Haverkos BM, Harris PJ. Investigational cancer drugs targeting cell metabolism in clinical development. Expert Opin Investig Drugs. 2015; 24(1):79–94.

13. Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927; 8(6):519–30.

16. Mankoff DA, Eary JF, Link JM, Muzi M, Rajendran JG, Spence AM, et al. Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond. Clin Cancer Res. 2007; 13(12):3460–9.

17. Weinhouse S. The Warburg hypothesis fifty years later. Z Krebsforsch Klin Onkol Cancer Res Clin Oncol. 1976; 87(2):115–26.

18. Jang M, Kim SS, Lee J. Cancer cell metabolism: implications for therapeutic targets. Exp Mol Med. 2013; 45:e45.

19. Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008; 13(6):472–82.

20. Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006; 441(7092):437–43.

21. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004; 4(11):891–9.

22. Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006; 66(2):632–7.

23. Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev. 2007; 26(2):299–310.

24. Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007; 109(9):3812–9.

25. Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013; 123(9):3678–84.

26. Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979; 254(8):2669–76.

27. DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007; 104(49):19345–50.

28. Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012; 491(7424):364–73.

29. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011; 11(2):85–95.

30. Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008; 105(48):18782–7.

31. David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010; 463(7279):364–8.

32. Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006; 312(5780):1650–3.

33. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006; 126(1):107–20.

34. Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012; 149(6):1269–83.

35. Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012; 3:e248.

36. Vicha A, Taieb D, Pacak K. Current views on cell metabolism in SDHx-related pheochromocytoma and paraganglioma. Endocr Relat Cancer. 2014; 21(3):R261–77.

37. Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002; 30(4):406–10.

38. Dunn WB. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys Biol. 2008; 5(1):011001.

39. Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J Pharm Biomed Anal. 2014; 87:1–11.

40. Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012; 13(4):263–9.
41. You L, Zhang B, Tang YJ. Application of stable isotope-assisted metabolomics for cell metabolism studies. Metabolites. 2014; 4(2):142–65.
Go to : 
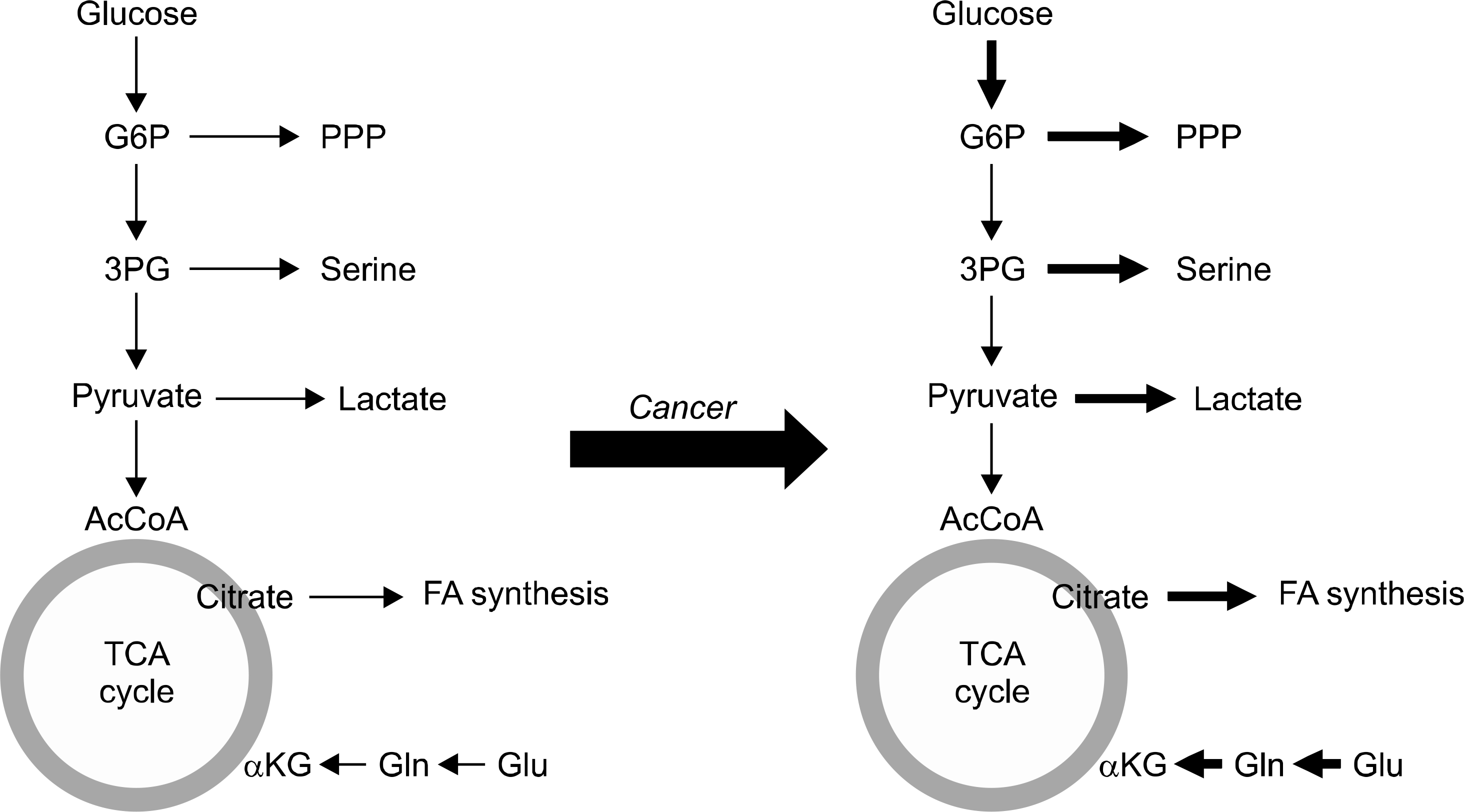 | Fig. 1.Summary of changes in metabolic characteristics of cancer cells. 3PG: 3-phos-phoglycerate, α KG: alpha- ketoglutarate, AcCoA: Acetyl coenzyme A, FA: fatty acids, G6P: glucose-6-phosphate, Gln: glutamate, Glu: gluta-mine, PPP: pentose phos-phate pathway. Bold arrow indicates increase of meta-bolic pathways in cancers. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download