Dear Editor,
Multidrug resistance of Pseudomonas aeruginosa has been attributed to both intrinsic and acquired antibiotic-resistance mechanisms. Multidrug-resistant (MDR) P. aeruginosa isolates have become a serious healthcare problem worldwide because they are resistant to almost all β-lactams, aminoglycosides, and quinolones. Production of zinc-dependent metallo-β-lactamases (MBLs) has been identified as the most significant mechanism among carbapenem-resistant P. aeruginosa isolates [1]. MBLs are of particular clinical concern because of their broad-spectrum activities, and Imipenemase (IMP)-, Verona Integron-Encoded Metallo-β-lactamase (VIM)-, Sao Paulo metallo-β-lactamase (SPM)-, Germany imipenemase (GIM)-, and New Delhi Metallo-β-lactamase (NDM)-type MBLs have been identified in P. aeruginosa worldwide [2]. Forty-six variants of VIM enzymes have been identified to date (http://www.lahey.org/Studies/other.asp). VIM-38 was recently identified in P. aeruginosa isolates in Turkey and was shown to differ from VIM-5 by a single substitution (Ala316Val) [3]. In P. aeruginosa, VIM-type MBLs have been reported within mobile genetic elements such as integrons, which contribute to the dissemination of antibiotic resistance [3].
We here report a new clinical P. aeruginosa strain isolated from a blood sample on January 2015 at Rize State Hospital in Turkey and identified by using the API 32GN system (bioMerieux, Marcy-l'Etoile, France). Minimal inhibitory concentrations were determined on a VITEK system for the following antibiotics: piperacillin/tazobactam, ceftazidime, cefepime, amikacin, netilmicin, ciprofloxacin, levofloxacin, imipenem, meropenem, cefoperazone-sulbactam, and inducible β-lactamase. 16S rDNA sequencing was used for molecular identification, performed according to Cicek et al [4].
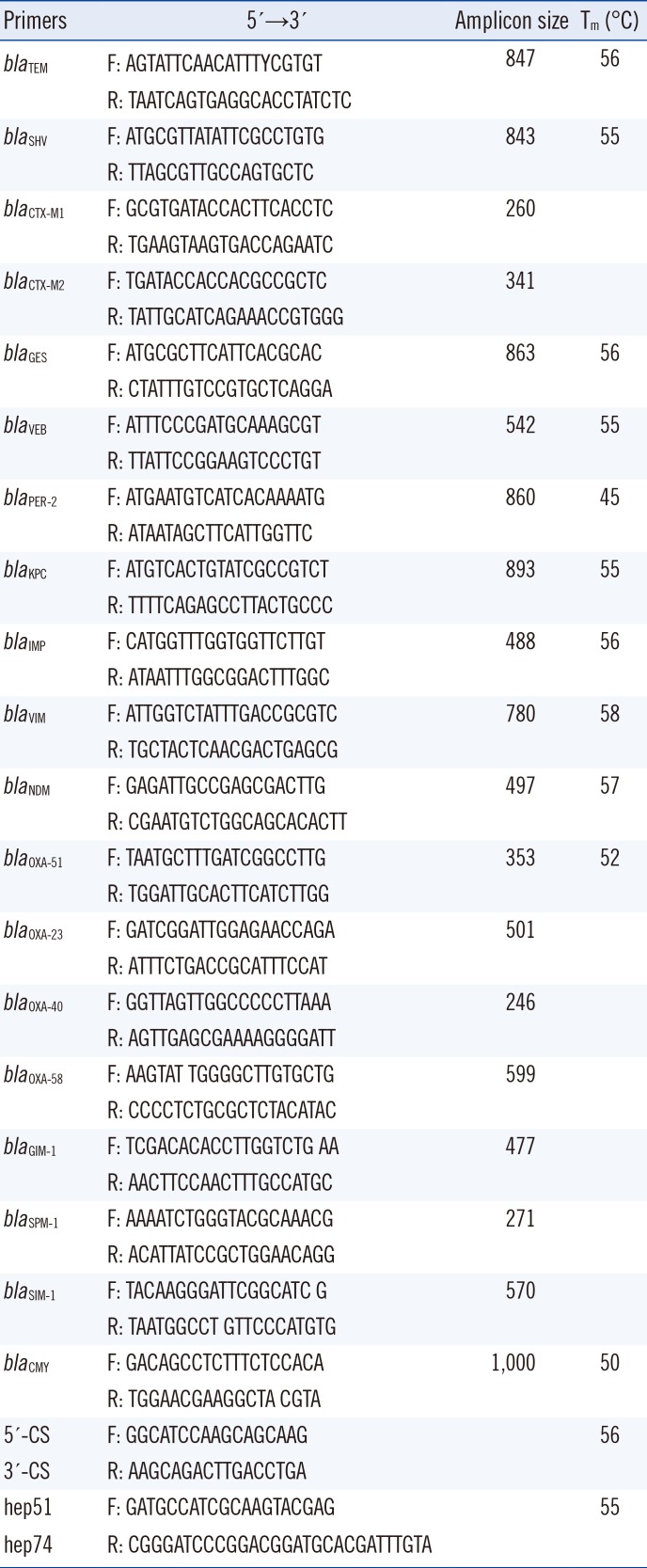
The P. aeruginosa isolate was screened for β-lactamase-encoding genes and the class 1–class 2 integrases conserved region by PCR. The primers used for detection of β-lactamase-encoding genes and class 1 and class 2 integron gene cassettes are listed in Table 1 [45678].
The positive PCR product of the class 1 integron was cloned into pGEM-T easy vector (Promega, Madison, WI, USA) and then sequenced by Macrogen (Amsterdam, The Netherlands). Sequencing results were analyzed by using the BLAST alignment search tool (http://www.ncbi.nlm.nih.gov/BLAST) and the multiple sequence alignment program CLUSTALW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Transferability of antibiotic resistance was tested according to the previously defined protocol [9], by using the rifampin-resistant Escherichia coli K-12 strain J53-2 as a recipient [4]. Susceptibility testing of the MBL-producing integron-positive P. aeruginosa isolate showed that it was resistant to imipenem, meropenem, piperacillin/tazobactam, ceftazidime, cefepime, and cefoperazone-sulbactam. PCR analysis showed that the isolate did not harbor any of the antibiotic resistance genes listed in Table 1, except for blaVIM-type MBL. Sequence analysis of the blaVIM-variant identified it as blaVIM-38. The P. aeruginosa isolate contained a class 1 integron gene cassette, but not a class 2 integron gene cassette. The blaVIM-38-harboring class 1 integron gene cassette was sequenced and was found to be 3,239 bp long. DNA sequence analysis revealed that blaVIM-38 MBL was located on the class 1 integron gene cassette together with AAC(6´)-Ib/EmrE/aadA1 (Fig. 1). The conjugation assay revealed that the class 1 integron cassette is not transferable.
The blaVIM-38 gene was identified in P. aeruginosa isolates in Turkey in 2014, and found to be located in a class 1 integron containing only two gene cassettes (blaVIM-38/orfD) [3]. This genetic structure has also been associated with the blaVIM-5 gene in a clinical isolate of Enterobacter cloaceae from Turkey [9]. Moreover, steady-state kinetic analyses in a study on the enzymatic properties of VIM-38 showed that VIM-38 hydrolyzed all of the tested penicillins, cephalosporins, and carbapenems [10].
In the present study, the class 1 integron included four gene cassettes with blaVIM-38 followed by AAC(6´)-Ib, EmrE (multi-drug transporter), and aadA1. This is the first report in Turkey of the blaVIM-38/AAC(6´)-Ib/EmrE/aadA1 gene cassette array. Therefore, we report a novel gene cassette array with an MBL gene in a P. aeruginosa clinical isolate, and this is the second report for the detection of VIM-38 in a P. aeruginosa isolate in Turkey with different hospitalization and isolation times.
In conclusion, the presence of class 1 integrons in P. aeruginosa leads to increased resistance to antibiotics. The present study demonstrates the emergence of VIM-producing MDR P. aeruginosa strains harboring class 1 integrons and a gene cassette in Turkey. In particular, the blaVIM-38 MBL gene appears to be spreading among P. aeruginosa isolates in Turkey.
Acknowledgments
This work was partially supported by Recep Tayyip Erdogan University Research Fund Grants (BAP-2014.102.03.03. and BAP-2014.102.03.02).
References
1. Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011; 11:381–393. PMID: 21530894.
2. Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007; 74:1686–1701. PMID: 17597585.
3. Iraz M, Duzgun AO, Cicek AC, Bonnin RA, Ceylan A, Saral A, et al. Characterization of novel VIM carbapenemase, VIM-38, and first detection of GES-5 carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa in Turkey. Diagn Microbiol Infect Dis. 2014; 78:292–294. PMID: 24428980.
4. Cicek AC, Duzgun AO, Saral A, Sandalli C. Determination of a novel integron-located variant (blaOXA-320) of Class D β-lactamase in Proteus mirabilis. J Basic Microbiol. 2014; 54:1030–1035. PMID: 24027220.
5. Iraz M, Özad Düzgün A, Sandallı C, Doymaz MZ, Akkoyunlu Y, Saral A, et al. Distribution of β-lactamase genes among carbapenem-resistant Klebsiella pneumoniae strains isolated from patients in Turkey. Ann Lab Med. 2015; 35:595–601. PMID: 26354347.
6. Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007; 59:321–322. PMID: 17185300.
7. Woodford N, Ellington MJ, Coelho JM, Turton JF, Warda ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006; 27:351–353. PMID: 16564159.
8. Zhao S, Qaiyumi S, Friedman S, Singh R, Foley SL, White DG, et al. Characterization of Salmonella enterica serotype newport isolated from humans and food animals. J Clin Microbiol. 2003; 41:5366–5371. PMID: 14662912.
9. Gacar GG, Midilli K, Kolayli F, Ergen K, Gundes S, Hosoglu S, et al. Genetic and enzymatic properties of metallo-β-lactamase VIM-5 from a clinical isolate of Enterobacter cloacae. Antimicrob Agents Chemother. 2005; 49:4400–4403. PMID: 16189133.
10. Makena A, Düzgün AÖ, Brem J, McDonough MA, Rydzik AM, Abboud MI, et al. Comparison of Verona Integron-Borne Metallo-β-lactamase (VIM) variants reveals differences in stability and inhibition profiles. Antimicrob Agents Chemother. 2015; 60:1377–1384. PMID: 26666919.
Fig. 1
Structure of the blaVIM-38-carrying class 1 integron gene cassette in Pseudomonas aeruginosa.
Abbreviations: Pint, integrase promoter; Pant, promoter of inserted gene(s); intI1, class 1 integrase; attI1, integron-associated recombination site; qacEΔ1, quaternary ammonium compound resistance gene cassette; ™, sulfonamide resistance gene; blaVIM-38, Verona Integron-Encoded Metallo-β-lactamase 38; CS, conserved segment.
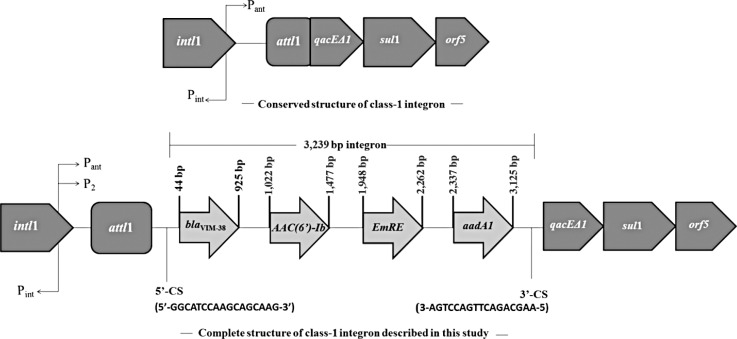
Table 1
Primers used in the amplification of selected genes




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download