Abstract
Objective
To determine the clinical significance of the polymerase chain reaction (PCR)-reverse dot blot (RDB) human papillomavirus (HPV) genotyping assay in cervical cancer screening.
Methods
A total of 10,442 women attending the Fujian Provincial Maternity and Children's Health Hospital were evaluated using the liquid-based cytology (thinprep cytologic test [TCT]) and the PCR-RDB HPV test. Women with HPV infection and/or abnormal cytology were referred for colposcopy and biopsy. For HPV DNA sequencing, 120 specimens were randomly selected. Pathological diagnosis was used as the gold standard.
Results
Using the PCR-RDB HPV test, overall HPV prevalence was 20.57% (2,148/10,442) and that of high-risk (HR)-HPV infection was 18.68% (1,951/10,442). There was 99.2% concordance between HPV PCR-RDB testing and sequencing. In this studied population, the most common HR-HPV types were HPV-16, −52, −58, −18, −53, −33, and −51, rank from high to low. HPV-16, −18, −58, −59, and −33 were the top 5 prevalent genotypes in cervical cancer but HPV-16, −18, −59, −45, and −33 were the top 5 highest risk factors for cancer (odds ratio [OR]=34.964, 7.278, 6.728, 6.101, and 3.658; all p<0.05, respectively). Among 10,442 cases, 1,278 had abnormal cytology results, of which, the HR-HPV positivity rate was 83.02% (1,061/1,278). To screen for cervical cancer by PCR-RDB HPV testing, when using CIN2+, CIN3+, and cancer as observed endpoints, the sensitivity was 90.43%, 92.61%, and 94.78% and the negative predictive value (NPV) was 99.06%, 99.42%, and 99.78%, respectively. PCR-RDB HPV and TCT co-testing achieved the highest sensitivity and NPV.
Go to : 
References
1. Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. Globocan 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012. Lyon: International Agency for Research on Cancer;2014.
2. Lewis DR, Chen HS, Midthune DN, Cronin KA, Krapcho MF, Feuer EJ. Early estimates of SEER cancer incidence for 2012: approaches, opportunities, and cautions for obtaining preliminary estimates of cancer incidence. Cancer. 2015; 121:2053–62.

3. Pan XF, Zhao ZM, Sun J, Chen F, Wen QL, Liu K, et al. Acceptability and correlates of primary and secondary prevention of cervical cancer among medical students in southwest China: implications for cancer education. PLoS One. 2014; 9:e110353.

4. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. 2007; 90:1–636.
5. Meijer CJ, Snijders PJ, Castle PE. Clinical utility of HPV genotyping. Gynecol Oncol. 2006; 103:12–7.

6. Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007; 121:621–32.

7. Wheeler CM, Hunt WC, Joste NE, Key CR, Quint WG, Castle PE. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst. 2009; 101:475–87.

8. AlObaid A, Al-Badawi IA, Al-Kadri H, Gopala K, Kandeil W, Quint W, et al. Human papillomavirus prevalence and type distribution among women attending routine gynecological examinations in Saudi Arabia. BMC Infect Dis. 2014; 14:643.

9. Zhang HY, Fei MD, Jiang Y, Fei QY, Qian H, Xu L, et al. The diversity of human papillomavirus infection among human immunodeficiency virus-infected women in Yunnan, China. Virol J. 2014; 11:202.

10. Moosa K, Alsayyad AS, Quint W, Gopala K, DeAntonio R. An epidemiological study assessing the prevalence of human papillomavirus types in women in the Kingdom of Bahrain. BMC Cancer. 2014; 14:905.

11. Poljak M, Marin IJ, Seme K, Vince A. Hybrid Capture II HPV Test detects at least 15 human papillomavirus genotypes not included in its current high-risk probe cocktail. J Clin Virol. 2002; 25(Suppl 3):S89–97.

12. Einstein MH, Martens MG, Garcia FA, Ferris DG, Mitchell AL, Day SP, et al. Clinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytology. Gynecol Oncol. 2010; 118:116–22.

13. Schutzbank TE, Jarvis C, Kahmann N, Lopez K, Weimer M, Yount A. Detection of high-risk papillomavirus DNA with commercial invader-technology-based analyte-specific reagents following automated extraction of DNA from cervical brushings in ThinPrep media. J Clin Microbiol. 2007; 45:4067–9.

14. Castle PE, Solomon D, Saslow D, Schiffman M. Predicting the effect of successful human papillomavirus vaccination on existing cervical cancer prevention programs in the United States. Cancer. 2008; 113:3031–5.

15. Day SP, Hudson A, Mast A, Sander T, Curtis M, Olson S, et al. Analytical performance of the investigational use only Cervista HPV HR test as determined by a multi-center study. J Clin Virol. 2009; 45(Suppl 1):S63–72.

16. Waxman AG, Chelmow D, Darragh TM, Lawson H, Moscicki AB. Revised terminology for cervical histopathology and its implications for management of high-grade squamous intraepithelial lesions of the cervix. Obstet Gynecol. 2012; 120:1465–71.

17. Pan QJ, Hu SY, Guo HQ, Zhang WH, Zhang X, Chen W, et al. Liquid-based cytology and human papillomavirus testing: a pooled analysis using the data from 13 population-based cervical cancer screening studies from China. Gynecol Oncol. 2014; 133:172–9.

18. Jun SY, Park ES, Kim J, Kang J, Lee JJ, Bae Y, et al. Comparison of the cobas 4800 HPV and HPV 9G DNA chip tests for detection of high-risk human papillomavirus in cervical specimens of women with consecutive positive HPV tests but negative Pap smears. PLoS One. 2015; 10:e0140336.

19. Abraham J, Stenger M. Cobas HPV test for first-line screening for cervical cancer. J Community Support Oncol. 2014; 12:156–7.

20. Huh WK, Ault KA, Chelmow D, Davey DD, Goulart RA, Garcia FA, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol. 2015; 136:178–82.

21. Li Y, Chen Z, Zhao L, Wang L, Tian M, Huang H, et al. Molecular diagnosis for a novel deletion mutation of α thalassemia. Zhonghua Xue Ye Xue Za Zhi. 2014; 35:724–7.
22. Hao Y, Xu ZY, Jin Q, Wu WQ, Cai J, Luo CQ, et al. Molecular and prenatal diagnosis for a Chinese pregnant woman with a novel mutation of β thalassemia. Zhonghua Xue Ye Xue Za Zhi. 2011; 32:245–8.
23. Yang G, Cui JH, Chen S, Si JH, Tan JJ, Li PY, et al. Establishment of a new HBV genotyping method with PCR-RBD and its application. Zhonghua Gan Zang Bing Za Zhi. 2004; 12:677–80.
24. Peng Q, Li S, Ma K, Li W, Ma Q, He X, et al. Large cohort screening of G6PD deficiency and the mutational spectrum in the Dongguan District in Southern China. PLoS One. 2015; 10:e0120683.

25. Pan QJ, Hu SY, Zhang X, Ci PW, Zhang WH, Guo HQ, et al. Pooled analysis of the performance of liquid-based cytology in population-based cervical cancer screening studies in China. Cancer Cytopathol. 2013; 121:473–82.

26. Belinson JL, Wu R, Belinson SE, Qu X, Yang B, Du H, et al. A population-based clinical trial comparing endocervical high-risk HPV testing using hybrid capture 2 and Cervista from the SHENCCAST II Study. Am J Clin Pathol. 2011; 135:790–5.

27. Wang Y, Yu YH, Shen K, Xiao L, Luan F, Mi XJ, et al. Cervical cancer screening and analysis of potential risk factors in 43,567 women in Zhongshan, China. Asian Pac J Cancer Prev. 2014; 15:671–6.

28. Stoler MH, Austin RM, Zhao C. Point-counterpoint: cervical cancer screening should be done by primary human papillomavirus testing with genotyping and reflex cytology for women over the age of 25 years. J Clin Microbiol. 2015; 53:2798–804.

30. Ishida K, Araki A, Kobayashi M, Taniyama K, Nabika T, Nagasaki M. An evaluation of the diagnostic and prognostic significance of p16 (INK4a)/p21 (WAF1/Cip1) immunostaining in squamous intraepithelial lesions of the uterine cervix using liquid-based cytology specimens. Diagn Cytopathol. 2014; 42:125–33.
31. Skinner SR, Wheeler CM, Romanowski B, Castellsagué X, Lazcano-Ponce E, Del Rosario-Raymundo MR, et al. Progression of HPV infection to detectable cervical lesions or clearance in adult women: analysis of the control arm of the VIVIANE study. Int J Cancer. 2016; 138:2428–38.

32. Katki HA, Schiffman M, Castle PE, Fetterman B, Poitras NE, Lorey T, et al. Five-year risks of CIN 3+ and cervical cancer among women who test Pap-negative but are HPV-positive. J Low Genit Tract Dis. 2013; 17:S56–63.
Go to : 
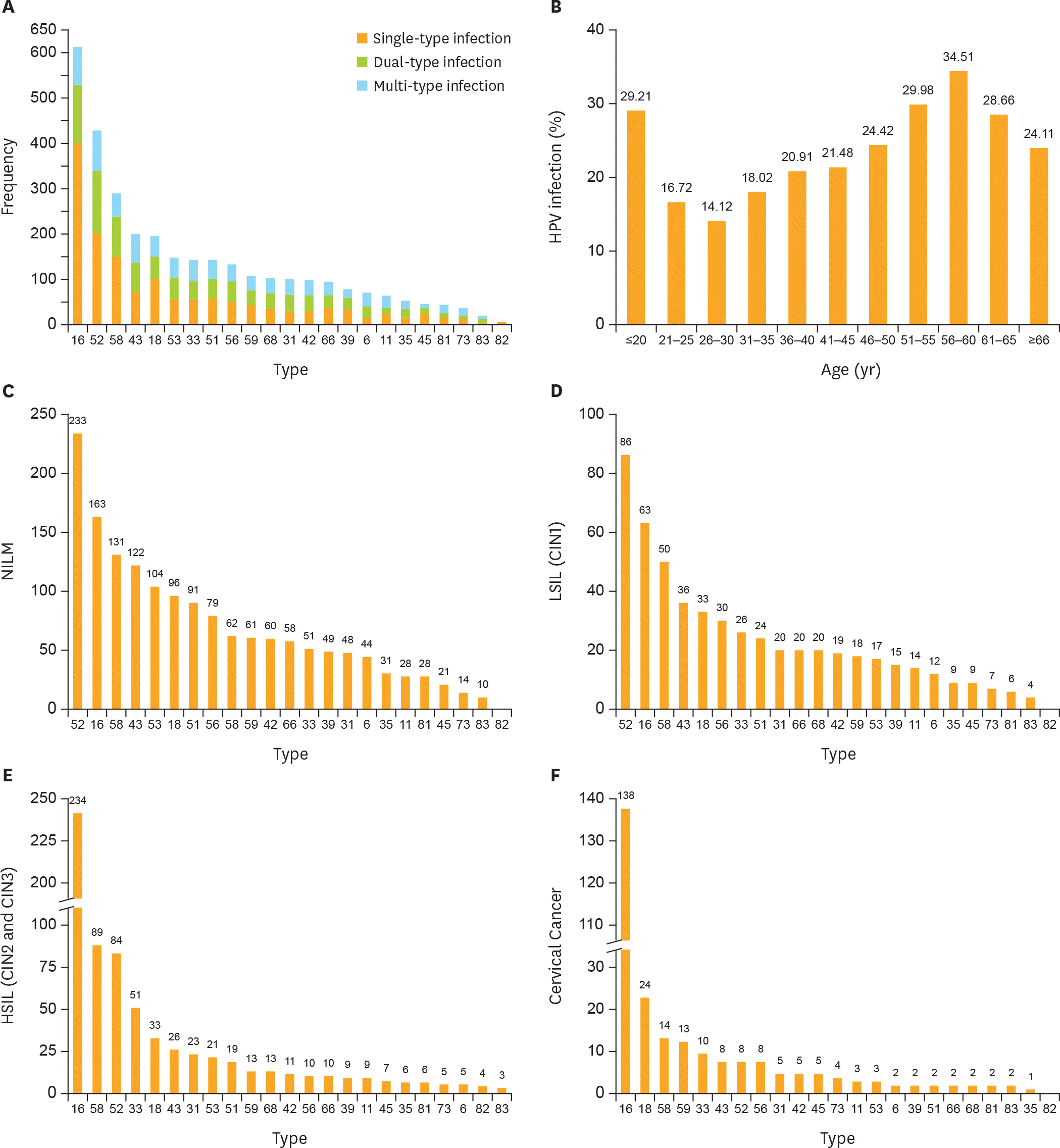 | Fig. 1.Prevalence of different HPV genotype infections. (A) HPV-16 is most prevalent genotype in HPV-positive women. (B) The HR-HPV infection rate showed a bimodal trend, and the rates of different age groups were statistically significant (χ2=272.740; p<0.001). Top 5 most frequent HPV genotypes in (C) NILM patients; (D) LSIL (CIN1) patients; (E) HSIL patients (CIN2 and CIN3); and (F) cervical cancer. Orange bar in (C-F) is the cumulative cases of HPV genotypes. CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; HR-HPV, high-risk human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy. |
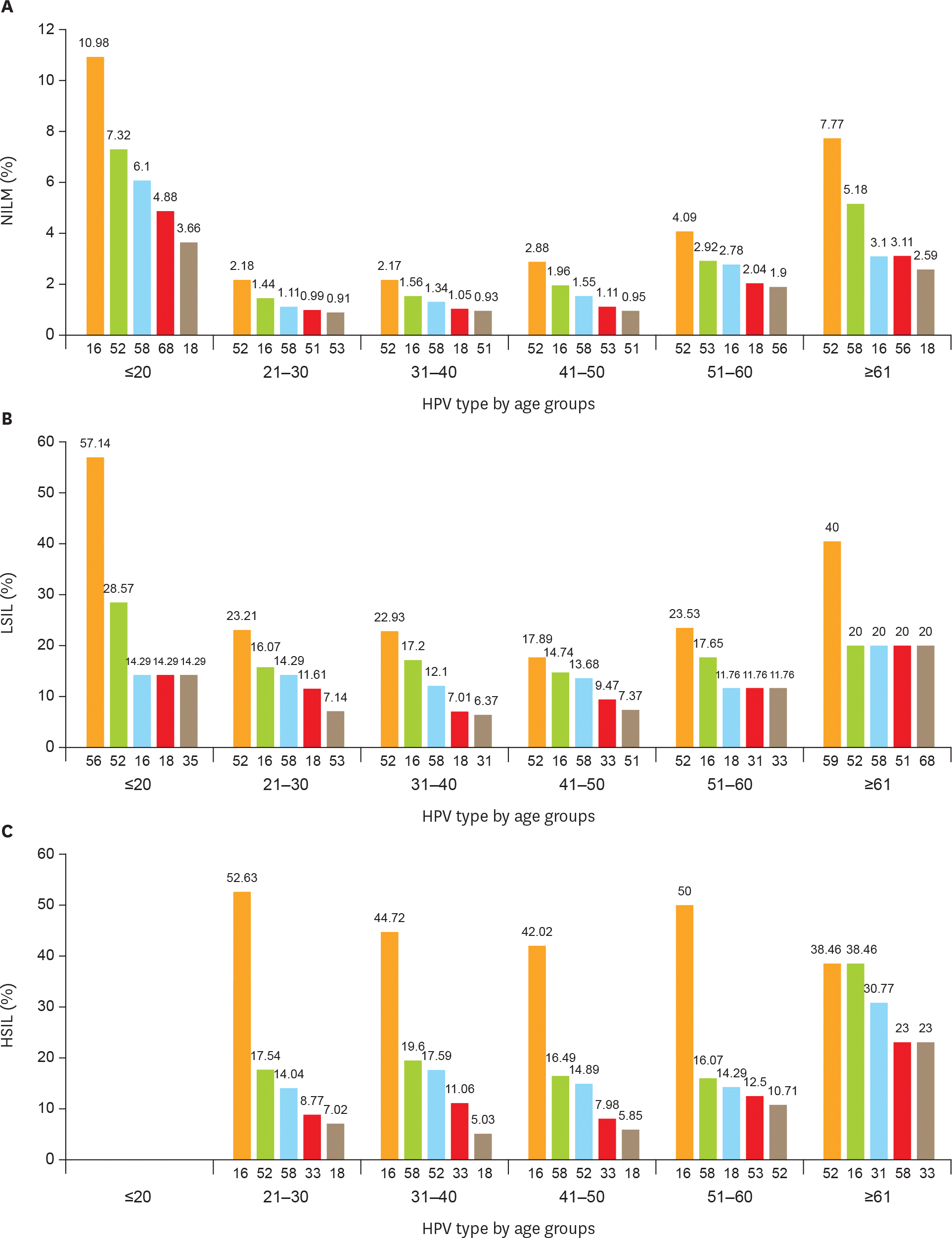 | Fig. 2.Age-related distribution of HPV infection. Top 5 most frequent HPV genotypes in different age groups of (A) NILM patients; (B) LSIL (CIN1) patients; (C) HSIL patients (CIN2 and CIN3); and (D) cervical cancer. CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy. (continued to the next page) |
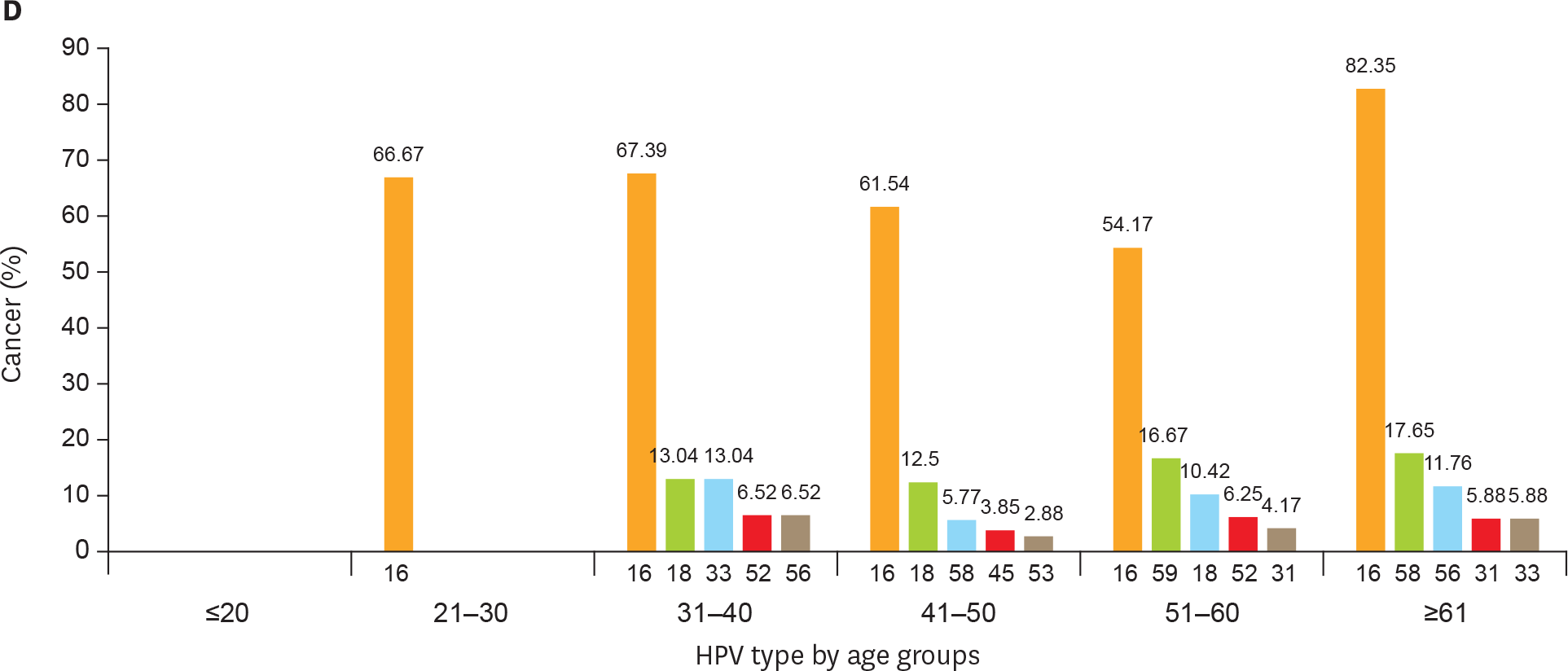 | Fig. 2.(Continued) Age-related distribution of HPV infection. Top 5 most frequent HPV genotypes in different age groups of (A) NILM patients; (B) LSIL (CIN1) patients; (C) HSIL patients (CIN2 and CIN3); and (D) cervical cancer. CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy. |
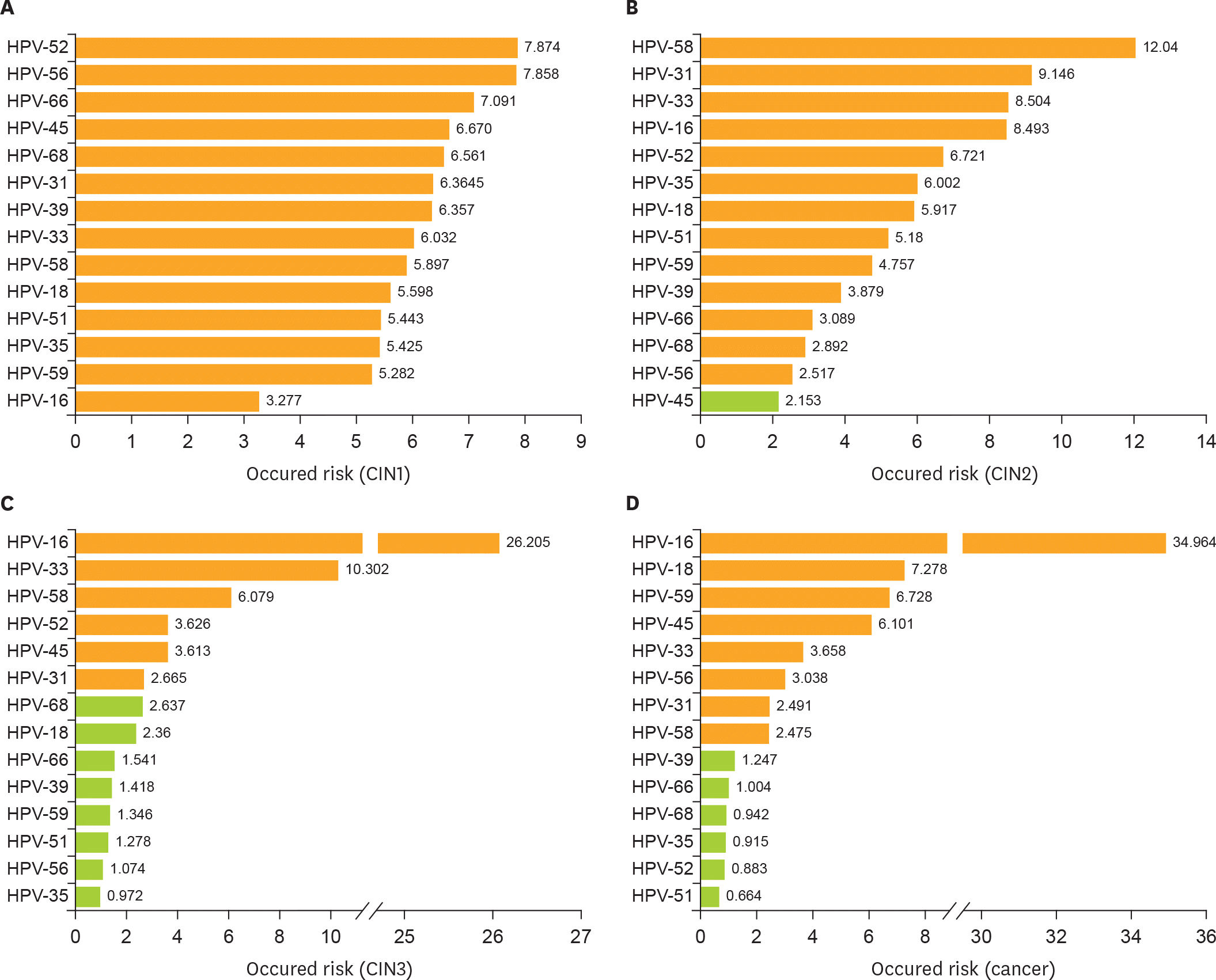 | Fig. 3.Cumulative occurred risk of HR-HPV genotype for cervical lesions. Cumulative occurred risk of each HPV genotype: in patients with (A) a pathologically diagnosed CIN1; (B) CIN2; (C) CIN3; and (D) invasive cervical cancer. Orange bar represents statistically significant (p<0.05) green bar represents not statistically significant (p>0.05). CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; HR-HPV, high-risk human papillomavirus. |
Table 1.
HPV infection by age related distribution (n=10,442)
| Age (yr) |
Negative (n=8,294) |
Single-type HPV (n=1,449) |
Dual-type HPV* (n=466) |
Multi-type HPV† (n=233) |
Total HPV infection (n=2,148)‡ |
|---|---|---|---|---|---|
| ≤20 (n=89) | 63 | 5 | 11 | 10 | 26 (29.21) |
| 21–25 (n=861) | 717 | 92 | 30 | 22 | 144 (16.72) |
| 26–30 (n=1,749) | 1,502 | 161 | 56 | 30 | 247 (14.12) |
| 31–35 (n=1,693) | 1,388 | 201 | 72 | 32 | 305 (18.02) |
| 36–40 (n=1,765) | 1,396 | 271 | 80 | 18 | 369 (20.91) |
| 41–45 (n=1,853) | 1,455 | 278 | 84 | 36 | 398 (21.48) |
| 46–50 (n=1,335) | 1,009 | 238 | 60 | 28 | 326 (24.42) |
| 51–55 (n=537) | 376 | 113 | 32 | 16 | 161 (29.98) |
| 56–60 (n=284) | 186 | 54 | 24 | 20 | 98 (34.51) |
| 61–65 (n=164) | 117 | 21 | 11 | 15 | 47 (28.66) |
| ≥66 (n=112) | 85 | 15 | 6 | 6 | 27 (24.11) |
Table 2.
Compare HPV infection and TCT with pathological diagnosis (n=10,442)
| Subjects | Pathologic diagnosis | χ2* | p-value | χ2† | p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| NILM (n=9,309) | CIN1 (n=395) | CIN2 (n=227) | CIN3 (n=292) | Cancer (n=219) | |||||
| TCT (n=10,442) | |||||||||
| HPV-positive (n=2,148) | 1,155 | 333 | 197 | 262 | 201 | ||||
| HPV-negative (n=8,294) | 8,154 | 62 | 30 | 30 | 18 | ||||
| Cytological diagnosis | |||||||||
| NILM (n=9,164) | 743.729 | 0.001 | 1,125.568 | 0.001 | |||||
| HPV-positive (n=1,087) | 890 | 63 | 52 | 49 | 33 | ||||
| HPV-negative (n=8,077) | 8,027 | 14 | 19 | 12 | 5 | ||||
| ASC (n=587) | 22.125 | 0.001 | 38.252 | 0.001 | |||||
| HPV-positive (n=437) | 168 | 142 | 41 | 57 | 29 | ||||
| HPV-negative (n=150) | 99 | 36 | 6 | 6 | 3 | ||||
| LSIL (n=312) | 0.584 | 0.540 | 3.103 | 0.524 | |||||
| HPV-positive (n=283) | 68 | 118 | 47 | 39 | 11 | ||||
| HPV-negative (n=29) | 6 | 11 | 4 | 5 | 3 | ||||
| ≥HSIL (n=317)‡ | 10.688 | 0.011 | 10.741 | 0.020 | |||||
| HPV-positive (n=300) | 10 | 4 | 53 | 111 | 122 | ||||
| HPV-negative (n=17) | 4 | 0 | 1 | 7 | 5 | ||||
| ≥AGC (n=62)§ | 5.866 | 0.019 | 9.103 | 0.038 | |||||
| HPV-positive (n=41) | 19 | 6 | 4 | 6 | 6 | ||||
| HPV-negative (n=21) | 18 | 1 | 0 | 0 | 2 | ||||
ASC, atypical squamous cells; AGC, atypical glandular cells; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy; TCT, thinprep cytologic test.
Table 3.
Compare cytology and HPV in the different degree of cervical lesion detection (n=10,442)
| Variable | Sensitivity | Specificity | PPV | NPV | PLR | NLR |
|---|---|---|---|---|---|---|
| CIN2+* | ||||||
| HPV | 90.43 (88.21–92.65) | 84.83 (84.12–85.55) | 31.02 (29.05–32.98) | 99.06 (98.85–99.27) | 5.8977 (5.5919–6.2202) | 0.1246 (0.1010–0.1537) |
| TCT | 76.96 (73.93–80.00) | 92.73 (92.21–93.25) | 44.65 (41.92–47.39) | 98.14 (97.86–98.42) | 10.5837 (9.7569–11.4807) | 0.2484 (0.2177–0.2835) |
| HPV+TCT | 95.26 (93.72–96.79) | 83.04 (82.29–83.79) | 29.98 (28.12–31.83) | 99.57 (99.42–99.71) | 5.6162 (5.3588–5.8861) | 0.0571 (0.0413–0.0789) |
| CIN3+† | ||||||
| HPV | 92.61 (89.08–93.64) | 83.20 (82.46–83.93) | 21.76 (20.00–23.51) | 99.42 (99.26–99.58) | 5.3918 (5.1188–5.6793) | 0.1129 (0.0862–0.1478) |
| TCT | 80.63 (77.20–84.05) | 91.32 (90.77–91.87) | 32.39 (29.82–34.96) | 98.92 (98.71–99.13) | 9.2889 (8.6029–10.0296) | 0.2122 (0.1777–0.2532) |
| HPV+TCT | 96.67 (95.12–98.23) | 81.32 (80.55–82.09) | 21.07 (19.75–22.72) | 99.79 (99.69–99.89) | 5.1747 (4.9514–5.4081) | 0.0409 (0.0256–0.0653) |
| Cervical cancer | ||||||
| HPV | 94.78 (91.14–95.42) | 81.11 (80.35–81.87) | 9.45 (8.27–10.69) | 99.78 (99.68–99.88) | 4.8581 (4.5915–5.1403) | 0.1013 (0.0651–0.1578) |
| TCT | 82.65 (77.63–87.66) | 89.30 (88.70–89.90) | 14.23 (12.31–16.15) | 99.58 (99.45–99.72) | 7.7270 (7.1142–8.3925) | 0.1943 (0.1455–0.2594) |
| HPV+TCT | 97.72 (95.74–99.70) | 79.11 (78.31–79.90) | 9.13 (8.65–10.29) | 99.94 (99.88–99.99) | 4.6772 (4.4810–4.8820) | 0.0289 (0.0121–0.0686) |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download