Abstract
Purpose
In order to suggest optimal anticancer drugs for patient-tailored chemotherapy, we developed a colorectal cancer (CRC)-liver metastasis patient-derived tumor xenograft (PDTX) model.
Methods
Tissue obtained from a patient with CRC-liver metastasis (F0) was transplanted in a nonobese female mouse with diabetic/severe combined immune deficiency (F1) and the tumor tissue was retransplanted into nude mice (F2). When tumor volumes reached ~500 mm3, the F2 mice were randomly divided into 4 groups (n = 4/group) of doxorubicin, cisplatin, docetaxel, and nontreated control groups. The tumor tissues were investigated using H&E staining, terminal deoxynucleotidyl transferase dUTP nick end labeling assays, and immunohistochemistry. To determine where the mutant allele frequencies varied across the different passages, we isolated genomic DNA from the primary tumor, liver metastasis, and PDTX models (F1/F2).
Results
The physiological properties of the tumor were in accord with those of the patient's tumors. Anticancer drugs delayed tumor growth, inhibited proliferation, and caused apoptosis. Histological assessments revealed no observable heterogeneity among the intragenerational PDTX models. Target exon sequencing analysis without high-quality filter conditions revealed some genetic variations in the 83 cancer-related genes across the generations. However, when de novo mutations were defined as a total count of zero in F0 and ≥5 in F2, exactly prognostic impact of clone cancer profiling (EGFR, KRAS, BRAF, PIK3CA, NRAS, APC and TP53) were detected in the paired.
Colorectal cancer (CRC) is one of the most common and lethal malignancies in Korea [1]. However, despite the recent progress in diagnostic tools that has allowed for early cancer diagnosis, tumor recurrence remains a major problem. Approximately 30% of recurrent CRC involves liver metastasis. Liver resection is not indicated in more than half of such recurrent cases. Recently, the survival rates in patients with liver metastasis from CRC have increased due to improvements in anticancer drugs. Two of the standard treatments for CRC-liver metastasis include combinations of 5-fluorouracil with oxaliplatin (FOLFOX) or irinotecan (FOLFIRI) [2]. Recently, a phase II study on FOLFOXIRI plus panitumumab was reported [3]. However, this particular regimen is not suitable for all patients with CRC-liver metastasis. Nevertheless, this initial chemotherapy plays an important role in converting unresectable CRC-liver metastases to resectable metastases [4]. Therefore, individualized chemotherapies might affect patient prognosis.
To determine chemotherapeutic efficacy, an animal model that preserves the physiological properties of patient tumors must be established. This task might be achieved using a patient-derived tumor xenograft (PDTX) model that maintains the histopathological architecture of the original tumor. Therefore, many researchers have attempted to develop a suitable PDTX model to predict therapeutic efficacy [567]. Such a model must offer a good platform for anticancer drug development and for monitoring drug response. Studies on this subject have increased over the past 10 years, especially for breast and lung cancer-derived xenograft models. Most developed PDTX models target primary cancers. Although metastatic disease leads to more mortality than primary cancer, comparatively little effort has been expended on developing xenograft models of metastasis.
The purpose of this study was to characterize the histological and genomic fidelities of CRC-liver metastasis PDTX models and to evaluate the application of CRC-liver metastasis PDTX models to the development of patient-tailored chemotherapy.
This collection of CRC-liver metastatic tissues was approved by the Institutional Review Board of Gangnam Severance Hospital (3-2014-0298). The samples were obtained from patients with pathologically proven CRC adenocarcinoma.
Female, nonobese diabetic (NOD)/severe combined immune deficiency (SCID) mice (5 to 8 weeks old) and female Balb/c nu/nu mice (5 weeks old) were purchased from SLC (Japan SLC Inc., Hamamatsu, Japan). All animal care and experiments were performed in accordance with the guidelines issued by the Institutional Animal Care and Use Committee in University.
Tissues were soaked in DMEM (GenDEPOT, Barker, TX, USA), transported on ice, and stored at 4℃ until transplantation. Tumor tissues were dissected into 1 mm × 1 mm fragments [8]. After an NOD/SCID mouse was anesthetized with isoflurane, 2 pieces of tumor specimens were implanted into the animal's flank. After the tumor volumes reached 500 mm3, the patientderived tumor tissues were obtained from the NOD/SCID mouse (the PDTX [F1] model). These isolated tissues were divided into several pieces, and 1 piece was fixed in 4% paraformaldehyde solution for histology and immunohistochemistry. The dissected pieces were re-engrafted into the flanks of Balb/c nu/nu mice (the PDTX [F2] model), while the remaining pieces were soaked in 5% dimethyl sulfide/fetal bovine serum and stored in liquid N2.
Tumor tissues were fixed in 4% paraformalin solution and embedded in paraffin. Paraffin blocks were sectioned into 5-µm slices. For histological observation, tissue sections were stained with hematoxylin and eosin (H&E). For immunohistochemistry, the tissue sections were deparaffinized in xylene, rehydrated with graded ethanol (from 100% to 70%), and treated with 3% H2O2 in methanol. The sections were then incubated with primary antibodies to Ki-67 (1:1000), CK7 (1:2000), and CK20 (1:500). All primary antibodies were obtained from DAKO (Carpinteria, CA, USA). After incubation with secondary antibodies, the tissue sections were treated with diaminobenzidine (DAB) solution and counterstained with hematoxylin.
Tumors were isolated from the NOD/SCID mouse (F1) and reimplanted into the flanks of nude mice. Two to 3 times per week, the tumor dimensions were measured using a caliper. Tumor volumes were calculated using the following formula:
To assess chemotherapeutic efficacy, nude mice (F2) were divided into 3 drug treatment groups and 1 control group (n = 4/group). Commercial drugs (cisplatin [CDDP], doxorubicin [DOX], and docetaxel [DTX], 10 mg/kg) were intravenously injected via a tail vein. Tumor sizes were measured 2–3 times per week. Body weights were recorded to evaluate toxicities.
After measuring tumor growth in F2 mice, the isolated tumor tissues were embedded in OCT compound (Leica Biosystems, Nussolch, Germany) and sectioned at 5 µm. Tissue sections were hydrated with 70% ethanol and treated with 3% H2O2 in methanol. The sections were then treated with TdT labeling buffer and incubated in TdT (Sigma-Aldrich, St. Louis, MO, USA)/biotinylated deoxyuridine (Roche Diagnostics, Mannheim, Germany). After reaction, the tissue sections were incubated with ABC complex and stained with DAB solution.
Genomic DNA was extracted from the primary tumor tissues and patient derived Xenograft (PDX) tissues using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacture's protocols. The DNA quality was assessed with 1% agarose gel electrophoresis and by the PicoGreen dsDNA Assay (Invitrogen, Carlsbad, CA, USA). The targeted panel was used to capture the target region covering 83 cancer-related genes. All of the coding exons of the following 72 genes were assessed for the detection of single-nucleotide variant (SNV), insertion/deletions (INDELs), and copy number variations (CNVs): ABL1, AKT1, AKT2, AKT3, ALK, APC, ARID1A, ARID1B, ARID2, ATM, AURKA, AURKB, BCL2, BRAF, BRCA1, BRCA2, CDH1, CDK4, CDK6, CDKN2A, CSF1R, CTNNB1, DDR2, EGFR, EPHB4, ERBB2, ERBB3, ERBB4, EWSR1, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, IDH2, IGF1R, ITK, JAK1, JAK2, JAK3, KDR, KIT, KRAS, MDM2, MET, MLH1, MPL, MTOR, NF1, NOTCH1, NPM1, NRAS, NTRK1, PDGFRA, PDGFRB, PIK3CA, PIK3R1, PTCH1, PTCH2, PTEN, PTPN11, RB1, RET, ROS1, SMAD4, SMARCB1, SMO, SRC, STK11, SYK, TERT, TMPRSS2, TOP1, TP53, and VHL. In addition, some introns of the following 5 genes were included in the detection of gene fusions: ALK, RET, ROS1, EWSR1, and TMPRSS2 (SureSelect, Agilent, Inc., USA). Approximately 200–500 ng of genomic DNA were prepared to construct libraries using the SureSelect targeted panel according to the manufacturer's protocol. Briefly, the qualified genomic DNA sample was randomly fragmented by Covaris followed by adapter ligation, purification, hybridization, and polymerase chain reaction. The captured libraries were subjected to the Agilent 2100 Bioanalyzer to estimate the quality and were loaded onto an Illumina HiSeq2500 (TheragenEtex Bio Institute, Suwon, Korea) according to the manufacturer's recommendations. The raw image files were processed by HCS1.4.8 for base-calling with default parameters. The sequences of each individual were generated as 101-bp paired-end reads.
At the NGS data pre-processing step, the sequence reads were aligned to the human genome (hg19) using BWA-MEM [9]. In order to generate the analysis-ready BAM, the overall pre-processing, including removal of duplication, local realignment, and recalibration, was performed using GATK Best Practice of Broad Institute [10].
At the variant discovery step, the SNVs and INDELs utilized 3 open source callers (UnifiedGenotyper [11], LoFreq [12], and SNVer [13]) and Samsung SDS's in-house callers. CNV and translocation were discovered with in-house callers developed by Samsung SDS. SNVs and INDELs were detected with an ensemble method that integrated three open source callers with an in-house caller. SNVs and INDELs were filtered using germ line mutations and false positive filters. SNVs with variant allele frequency (VAF) ≥ 5% and INDELs ≥ 10% were selected as the final results. CNVs between the tumor and preprocessed normal data were analyzed using the depth of coverage for each target region. To calculate the absolute copy number, tumor purity and ploidy were estimated from a statistical model using log2 ratio values and SNV VAF values. With regard to cutoff values, copy number (CN) ≥ 7 and CN = 0 were used for amplification and homo deletion, respectively. A paired-end mapping analysis and a split-alignment analysis were applied for translocation detection. All discordant read-pairs with an abnormal insert size or orientation were screened. Soft-clipping information of the split-reads was investigated as evidence of the genomic rearrangements. The cutoff value of the confident translocations was a split-read support count ≥ 3.
We collected surgically resected tumor samples from two patients with CRC liver metastasis and obtained only one patient PDTX model. As shown in Fig. 1A, the CRC-liver metastasis patient-derived tumors were transplanted into an NOD/SCID mouse (the PDTX [F1] model) and re-transplanted into nude mice (the PDTX [F2] model). When 2 pieces of the patientderived tumor tissue were transplanted into an NOD/SCID mouse the day after surgery, the mean latency period required to establish the PDTX model (F1) was 60 days. As shown in Fig. 1B, the tumor growth was faster in PDTX (F2) than it was in PDTX (F1). Therefore, we investigated the histological morphology and tissue characterization across the generations (F0, F1, and F2). Morphologies of F1 and F2 tissues resembled the original patient-derived colon cancer tissues (Fig. 2A). To confirm the histological characters of F2, we examined the markers of CRC-liver metastasis [8]. In F2 tissues, CK20 (a positive marker of metastasis) was detected. In contrast, CK7 (a negative marker of metastasis) was not detected in F2 tissues (Fig. 2B). These results indicate that F2 preserved the physiological properties of the original patient tumors.
To determine where the mutant allele frequencies varied across the different passages, we isolated genomic DNA from the primary tumor, liver metastasis, and PDTX models (F1/F2). Histological assessments reviewed no observable heterogeneity among the intragenerational PDTX models. Target exon sequencing analysis without high-quality filter conditions revealed some genetic variations in the 83 cancer-related genes across the generations (Table 1A). However, when de novo mutations were defined as a total count of zero in F0 and ≥5 in F3, exactly prognostic impact of clone cancer profiling (EGFR, KRAS, BRAF, PIK3CA, NRAS, APC and TP53) were detected in the paired tumors (Table 1B, C). This result suggests that the mutational changes in PDTX were preserved in the early passages in mice.
The chemotherapeutic efficacies of commercial anticancer agents were evaluated using the PDTX model. To screen the responses to anticancer drugs, we selected three anticancer drugs that act via different mechanisms. Fortunately, single treatments with all 3 drugs suppressed tumor growth in the F2 model. As shown in Fig. 3A, tumor proliferation was delayed by CDDP, DOX, and DTX. The doubling time of the tumors in the control group was 12 days. However, CDDP, DOX, and DTX delayed the doubling time by over 1 week. The CDDP-treated animals showed evidence of slight recurrence after 3 weeks. This result was presumably due to individual administration of these anticancer drugs. There was no body weight loss detected in any of the 4 groups (Fig. 3B).
To assess the therapeutic efficacy of these anticancer drugs, Ki-67 protein expression levels (a marker of proliferation) were measured (Fig. 4A). The Ki-67 expression levels (expressed as a percentage of brown spots) were as follows: 26.6% ± 3.9% in the control group, 24.7% ± 2.8% in the CDDP group, 21.0% ± 2.8% in the DOX group, and 19.0% ± 3.4% in the DTX group (95% confidence interval). These findings concurred with the observed tumor growth delay. We also investigated drug-induced apoptosis and necrosis. As shown in Fig. 4B, brown spots represented apoptotic cell death. The control group did not demonstrate any apoptotic cell death. In contrast, apoptotic cell death increased in the experimental groups in the order CDDP → DOX → DTX. Apoptosis induction is related to suppression of the proliferation marker level. Our results indicate that the anticancer drugs inhibited the proliferation of CRC-liver metastatic cancer cells and triggered their apoptosis. Necrosis was also observed. Gland-substituted foamy histiocytes reflect necrotic cell death. Therefore, we considered the gland-substituted foamy histiocytes as a surrogate of necrosis and calculated the ratio of necrotic glands to total glands. As shown in Fig. 4C, this ratio was <10% in the anticancer agent-treated groups (CDDP 6%, DOX 7%, DTX 8%, respectively). However, the ratio was 0% in the control group.
The aim of this study was to evaluate PDTX models to predict the response to chemotherapy. Although we attempted to create 2 models, we only successfully established one CRC-liver metastasis PDTX model. The metastatic lesions had a higher success rate than did the primary tumors [1415].
An evaluation process is needed to suggest chemotherapy guidelines based on the sensitivity of CRC-liver metastatic tissues to candidate drugs. Recently, techniques have been devised for personalized chemotherapy. The PDTX model, which preserves the physiological properties of patient tumors, is one such technique. This model is considered to be a good platform for chemosensitivity screening. Therefore, in this study, we established a PDTX model using CRC-liver metastatic cancer tissues.
Previous studies have demonstrated that tumorigenesis from patient-derived tumor tissues is slower than that from primary cancer cells or established cancer cell lines [16]. The rate of tumorigenesis also depends on the transplanted site (e.g., orthotopic or ectopic) and transplantation method [171819]. In this study, we attempted to establish a PDTX model and to ensure tumorigenesis using patient-derived tumor tissues in NOD/SCID mice using fresh tissue rather than cryopreserved tissue. It took 60 days to establish the PDTX F1 model, but less time to establish the F2 model.
The biggest advantage of PDTX models is that they preserve the genetic features and microenvironment of a patient's tumor. The histology of the primary tumor was shown to be maintained through successive generations in colon cancer [8], which is consistent with the results of our study. However, some genes were newly identified or disappeared in the PDTX models compared to those of the primary tumors, as in a previous report [20]. It is possible that heterogeneity in the primary tumor was a result of some mutations being detected in the primary tumor but not in PDTX, or vice versa. Alternatively, selection pressures can arise during engraftment into different species [20]. Regardless, we demonstrated that well-known cancer-related genes are preserved between primary tumor and PDXT models. Therefore, the PDTX models could constitute preclinical models that preserve the tumor microenvironment of the original patient.
We evaluated the efficacies of CDDP, DOX, and DTX. Their chemosensitivities were compared using tumor growth curves. The histological results reflected the modes of cell death responsible for tumor suppression. In this study, with regard to each drug, the inhibition rates in the PDTX models were similar to the tumor volume reduction. We concluded that the CRC-liver metastasis PDTX models could be used to deduce information regarding the sensitivity to therapeutic agent.
This study has several limitations. First, evaluations of the therapeutic agents are not ideal given that they were tested in immunocompromised mice. Another limitation is that all of the PDTX models were established in subcutaneous tissues. Prior evidence has suggested that the anatomical site can affect tumor biology and its response to chemotherapies. A third limitation is that the success rate varied significantly between patient tumor characteristics.
In conclusion, the CRC liver metastasis PDTX models developed in this study retain the genomic and histological characteristics of the original patient tumors. By integrating the predictive value of these models into therapeutic strategies, they may be used to develop personalized medicine.
Figures and Tables
 | Fig. 1Establishment of a patient-derived colorectal cancer-liver metastasis xenograft model. (A) Schematic of the patientderived tumor xenograft (PDTX) model. (B) Tumor growth PDTX model (F2). Tumor tissues were retransplanted into Balb/c nu/nu mice (n = 18). Results are presented as mean ± standard deviation. |
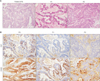 | Fig. 2Histologic evaluation of a patient-derived colorectal cancer-liver metastasis xenograft model. (A) Histology of a patient tumor and of the patient-derived tumor xenograft F1- and F2-derived tumors. Tissue sections were stained with H&E (×400). (B) Identification of the metastasis model. Tissue sections were immunostained for CK7 and CK20 (×200). |
 | Fig. 3Chemotherapeutic effects of anticancer drugs in the patient-derived tumor xenograft model. (A) Tumor growth delay of anticancer drugs. (B) Mouse body weights. Results are presented as mean ± standard deviation (n = 4). CDDP, cisplatin; DOX, doxorubicin; DTX, docetaxel. |
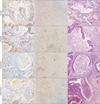 | Fig. 4Characterization of tumor tissues after treatment with anticancer drugs. (A) Immunostaining with Ki-67 as a marker of proliferation. Tissue sections were stained with hematoxylin (×200). (B) Tumor tissue apoptosis was quantified using the TUNEL assay. Brown dots represent apoptotic cells. Tissues were observed under a microscope (×400). (C) Tumor necrosis. Tissue sections were stained with H&E (×200). |
ACKNOWLEDGEMENTS
This research was supported by Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2016R1A6A1A03007648) and a grant from NRF (2014R1A1A3049498).
References
1. Jung KW, Won YJ, Kong HJ, Oh CM, Cho H, Lee DH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat. 2015; 47:127–141.
2. Puig I, Chicote I, Tenbaum SP, Arques O, Herance JR, Gispert JD, et al. A personalized preclinical model to evaluate the metastatic potential of patient-derived colon cancer initiating cells. Clin Cancer Res. 2013; 19:6787–6801.
3. Bendell JC, Zakari A, Peyton JD, Boccia R, Moskowitz M, Gian V, et al. A Phase II Study of FOLFOXIRI Plus Panitumumab Followed by Evaluation for Resection in Patients With Metastatic KRAS Wild-Type Colorectal Cancer With Liver Metastases Only. Oncologist. 2016; 21:279–280.
4. Akgul O, Cetinkaya E, Ersoz S, Tez M. Role of surgery in colorectal cancer liver metastases. World J Gastroenterol. 2014; 20:6113–6122.
5. Whittle JR, Lewis MT, Lindeman GJ, Visvader JE. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015; 17:17.
6. Izumchenko E, Meir J, Bedi A, Wysocki PT, Hoque MO, Sidransky D. Patient-derived xenografts as tools in pharmaceutical development. Clin Pharmacol Ther. 2016; 99:612–621.
7. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015; 21:1318–1325.
8. Cho YB, Hong HK, Choi YL, Oh E, Joo KM, Jin J, et al. Colorectal cancer patient-derived xenografted tumors maintain characteristic features of the original tumors. J Surg Res. 2014; 187:502–509.
9. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760.
10. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43:491–498.
11. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.10.1–11.10.33.
12. Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201.
13. Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011; 39:e132.
14. Nemati F, Sastre-Garau X, Laurent C, Couturier J, Mariani P, Desjardins L, et al. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin Cancer Res. 2010; 16:2352–2362.
15. Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, et al. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci Transl Med. 2012; 4:159ra149.
16. Boone JD, Dobbin ZC, Straughn JM Jr, Buchsbaum DJ. Ovarian and cervical cancer patient derived xenografts: The past, present, and future. Gynecol Oncol. 2015; 138:486–491.
17. Dong X, Guan J, English JC, Flint J, Yee J, Evans K, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res. 2010; 16:1442–1451.
18. Mohseni MJ, Amanpour S, Muhammadnejad S, Sabetkish S, Muhammadnejad A, Heidari R, et al. Establishment of a patient-derived Wilms’ tumor xenograft model: a promising tool for individualized cancer therapy. J Pediatr Urol. 2014; 10:123–129.
19. Zhu Y, Tian T, Li Z, Tang Z, Wang L, Wu J, et al. Establishment and characterization of patient-derived tumor xenograft using gastroscopic biopsies in gastric cancer. Sci Rep. 2015; 5:8542.
20. Hao C, Wang L, Peng S, Cao M, Li H, Hu J, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015; 357:179–185.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download