Abstract
Purpose
We evaluated the clinical role of rapid next-generation sequencing (NGS) for identifying BRCA1/2 mutations compared to traditional Sanger sequencing.
Methods
Twenty-four paired samples from 12 patients were analyzed in this prospective study to compare the performance of NGS to the Sanger method. Both NGS and Sanger sequencing were performed in 2 different laboratories using blood samples from patients with breast cancer. We then analyzed the accuracy of NGS in terms of variant calling and determining concordance rates of BRCA1/2 mutation detection.
Results
The overall concordance rate of BRCA1/2 mutation identification was 100%. Variants of unknown significance (VUS) were reported in two cases of BRCA1 and 3 cases of BRCA2 after Sanger sequencing, whereas NGS reported only 1 case of BRCA1 VUS, likely due to differences in reference databases used for mutation identification. The median turnaround time of Sanger sequencing was 22 days (range, 14–26 days), while the median time of NGS was only 6 days (range, 3–21 days).
Conclusion
NGS yielded comparably accurate results to Sanger sequencing and in a much shorter time with respect to BRCA1/2 mutation identification. The shorter turnaround time and higher accuracy of NGS may help clinicians make more timely and informed decisions regarding surgery or neoadjuvant chemotherapy in patients with breast cancer.
BRCA1 and BRCA2 (BRCA1/2) are tumor suppressor genes that play a crucial role in the development of hereditary breast and ovarian cancer (HBOC) syndrome [1]. Testing for BRCA1/2 mutations in patients with breast cancer is important because knowledge of these mutations helps clinicians to make a decision about various options of risk-reduction strategies [23]. The timing of preventive surgery partly depends on the timing of genetic testing and a patient's age [4]. For example, women who know their carrier status before or near the time of their breast cancer diagnosis can incorporate this information into decisions regarding primary surgical treatment [5]. It is necessary for patients with a strong family history of breast and ovarian cancer or high risk (>10%) of BRCA1/2 mutations to guide them through their choice of preventive surgery, if elected [6].
Traditional Sanger sequencing has a high cost and long turnaround time (TAT). In the United States, where commercial DNA testing is available, results are often reported within 2 weeks, but in Western Europe, Sanger sequencing TAT can reach 4 weeks [3]. However, because of low insurance coverage and the labor-intensive and time-consuming work-flow of Sanger sequencing analysis for BRCA1/2, a 4- to 6-week TAT is typical in South Korea; a 2-week TAT is not generally available in South Korea. As a result, most Korean patients newly diagnosed with breast cancer have undergone surgery or chemotherapy without knowledge of their BRCA1/2 genotype.
Next-generation sequencing (NGS) is a revolutionary high-throughput nucleotide sequencing method that delivers fast, inexpensive, and accurate genomic data [7]. This method can provide similar genetic information for clinicians at a lower cost and shorter TAT compared to Sanger sequencing [8]. Therefore, it is necessary to elucidate how NGS can be applied and what effects on it will have in clinical practice. However, before adopting NGS technology for identifying BRCA1/2 mutations, validation with traditional Sanger sequencing, the standard method of detecting these mutations, should be evaluated. In this study, we investigated the performance and diagnostic accuracy of NGS compared to Sanger sequencing in detecting BRCA1/2 mutations and the role of NGS in clinical practice for patients with breast cancer who are candidates for risk-reduction strategies.
To evaluate NGS validation as a modality of clinical mutation detection compared to Sanger sequencing, blood samples from 12 patients were used. All patients were prospectively enrolled in Severance Hospital between March 2014 and July 2014 and had been diagnosed with invasive breast cancer (Fig. 1). Patients' clinicopathologic features were summarized in Table 1. Written informed consent was obtained before blood sampling. Selection criteria for BRCA1/2 screening were based on the KOHBRA (Korean Hereditary Breast Cancer) study, which is covered by the national insurance system [9]. Both Sanger and NGS methods were simultaneously performed in 2 different laboratories in Korea. All genetic interpretations of each sample were independently performed in both laboratories, and sequencing results were not shared between laboratories. Sanger sequencing was performed by the clinical laboratory, SCL (Seoul, Korea), which is certified by the College of American Pathologists laboratory accreditation program. NGS was carried out in the clinical genetics laboratory in the Department of Laboratory Medicine at Severance Hospital. One of 12 patients who agreed to participate in the ongoing international randomized clinical trial received additional BRACAnalysis services (Myriad Genetics Inc., Salt Lake City, UT, USA). Because NGS was not fully validated by traditional Sanger sequencing at the beginning of the study and was not covered by the Korean national insurance system, genetic counseling and clinical decision-making for patients regarding their genetic results were accomplished based on traditional Sanger sequencing or BRACAnalysis.
Genomic DNA was extracted immediately upon arrival of the peripheral blood samples at the laboratory using the QIAamp DNA Mini Kit (QIAGEN, Victoria, Australia) on a QIAcube system (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The DNA quality was confirmed using a Qubit dsDNA HS Assay kit (Life Technologies, Carlsbad, CA, USA) on a Qubit2.0 Fluorometer (Life Technologies). If the samples were received within 2 days of each other, the 2 samples were run simultaneously. Library preparation was performed using the Ion AmpliSeq BRCA1 and BRCA2 Panel primer set, Ion AmpliSeq Library Kit 2.0, and Ion Xpress Barcode Adapters (Life Technologies) under the following conditions: (1) enzyme activation at 99℃ for 2 minutes, (2) denaturation at 99℃ for 15 seconds, (3) annealing and extension at 60℃ for 4 minutes (19 cycles), and (4) holding at 10℃. Purification was done using the Agencourt AMPure XP reagent (Beckman Coulter, Brea, CA, USA) and 70% ethanol on a DynaMagTM-96 Side Magnet (Life Technologies). Quality control for the amplicons was established with the High-Sensitivity DNA kit on the Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA). Multiplexing was accomplished by combining 100 pmol of amplicons into each library tube. Template preparation and emulsion polymerase chain reaction (PCR) was performed using the Ion Onetouch 200 template Kit v2 on the Ion OneTouch 2 system with Dynabeads MyOne Streptavidin C1 beads on the Ion OneTouch ES (Life Technologies). The spherical particles were sequenced on an Ion 314 Chip (Life Technologies) using a Ion PGM Sequencing 200 Kit v2 on the Ion PGM system (Life Technologies).
Ion Torrent reads were initially analyzed for variant detection using the Ion Torrent Variant Caller, which is available at the Life Technology Torrent Browser Plugin Store. Visual confirmation of the identified variants was accomplished with Integrated Genomics Viewer software from Broad Institute (Cambridge, MA, USA; http://www.broadinstitute.org/igv/). Further data review of missense variants was performed by using online software for predicting alterations of protein function, such as SIFT (Sorting Intolerant From Tolerant; http://sift.bii.a-star.edu.sg/index.html) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml). Filtering of the data was done manually by reviewing all variants identified by the Ion Torrent Variant Caller.
Genomic DNA was isolated using the QIAamp DNA Mini Kit. Sanger sequencing was performed as follows. Briefly, PCR amplification was performed using F-Taq polymerase (Solgent, Korea). Each 25-µL reaction contained 1X PCR buffer, 1.5-mmol/L MgCl2, 2 mmol/L of each dNTP, 5 pmol/L each of the forward and reverse primers, 0.5 U F-Taq polymerase, and 100-ng genomic DNA. The thermal cycling program included the follow steps: (1) 94℃ for 5 minutes, (2) 94℃ for 30 seconds, (3) appropriate annealing temperature for 30 seconds, (4) 72℃ for 45 seconds, and (5) 72℃ for 3 minutes. Steps 2–4 were repeated for 30 cycles. Primers were synthesized according to published primer sequences when available or were custom-designed [10]. Full coding regions of the BRCA1 and BRCA2 genes were amplified using single-locus conventional PCR. The targeted region was defined as the complete coding regions of BRCA1 and BRCA2 and approximately 20 bp of noncoding DNA flanking the 5′ and 3′ ends of each exon. Each PCR amplicon was treated with 20-µL reaction mixture comprising 3 U exonuclease I, 5X exonuclease I buffer, and 1.7 U FastAP thermosensitive alkaline phosphatase (Fermentas, Waltham, Massachusetts, USA) and incubated at 37℃ for 45 minutes, followed by heat-inactivation at 80℃ for 10 minutes. Cycle sequencing was performed using the BigDye Terminator kit v1.1 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Sequencing products were analyzed on a 3130xl Genetic Analyzer (Applied Biosystems). The SeqScape software v2.7 (Applied Biosystems) was used for visualization and sequence alignment of Sanger data.
References used for mutation identification by Sanger sequencing were Breast Cancer Information Core database (BIC; http://research.nhgri.nih.gov/bic/) and the Human Genome Mutation Database (HGMD; http://www.hgmd.org). Reporting of Sanger sequencing was performed using guidelines for mutation nomenclature of the Human Genome Variation Society (http://www.hgvs.org). References used for NGS analysis were based on the BIC, HGMD, and Database of Single Nucleotide Polymorphisms (dbSNP; http://www.ncbi.nlm.nih.gov/SNP/). Significant mutations were considered “positive” for BRCA1/2 mutations, and variants of unknown significance (VUS) and nonsignificant variants were considered “negative.”
Comparison of the continuous variable (TAT) was performed using the Student t-test. Numbers of mutation and VUS were compared using the chi-square test. All tests were 2-sided, and P-values less than 0.05 indicated statistical significance. Statistical analysis was performed using IBM SPSS Statistics ver. 20.0 (IBM Co., Armonk, NY, USA).
The authors confirm that all ongoing and related trials for this intervention are registered. All patients agreed with enrollment of the study. They signed informed consent forms before enrollment. This study was approved by the Severance Hospital Institutional Review Board (2013-1411-001), and the ClinicalTrials.gov identifier for this study is NCT02151747.
The median age of patients in our study was 36 years (range, 32–53 years), and 7 patients were less than 40 years old. Ten patients had primary breast cancer, 2 patients had metastatic breast cancer, and bilateral breast cancer was manifested in 2 patients. Three patients had a second-degree family history of breast or ovarian cancer.
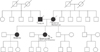
BRCA1/2 mutations were identified in 2 patients (2 of 12, 16.6%). Both Sanger sequencing and NGS identified one BRCA1 and one BRCA2 mutation carrier; thus, the overall accuracy rate of NGS was 100% (specificity 100% and sensitivity 100%). An example of the pedigree charts for a patient with BRCA1 mutation was illustrated in Fig. 2. However, among 10 patients whose Sanger and NGS analyses yielded negative BRCA1/2 mutation results, 1 patient received additional BRACAnalysis that included large genomic rearrangement test. This additional analysis revealed a deleterious mutation that was not detected by either Sanger sequencing or NGS.
Sanger sequencing identified 2 patients with BRCA1 VUS and 3 patients with BRCA2 VUS, although NGS reported only 1 patient with BRCA1 VUS. The reported VUS from Sanger sequencing were c.5590G>A, c.4883T>C, c.10234A>G, c.10234A>G, c.671-8A>G, and c.811G>A. The reported VUS from NGS were c.671-8A>G and c.811G>A in a patient less than 40 years old with metastatic breast cancer, which were also detected in this patient by Sanger sequencing. Differences in BRCA1/2 variants identification between the two sequencing methods were not statistically different (BRCA1, P=1.00; BRCA2, P=0.21) (Table 2).
Nonsignificant variants, including some synonymous and missense variants, were detected with NGS more often than with Sanger sequencing. The nonsignificant variants detected only by NGS were c.2510A>G (zygosity=19%), c.1961delA (zygosity=19%), c.68-4A>T (zygosity=17%), c1804G>A (zygosity=23%), c.3860delA, c8926delA (zygosity=32%), c.8941G>A (zygosity=22%), c.8942A>G (zygosity=14%), c.4563A>G, c.6513G>C, and c.7397T>C. Three variants identified by NGS (c.4563A>G, c.6513G>C, and c.7397T>C were found in all 12 patients as homozygous synonymous or missense variants of BRCA2. However, these three variants were not detected as variants of BRCA2 as reported by Sanger sequencing. Variants called with an inadequate zygosity possibility (zygosity<40–60%) using NGS disappeared after retesting. The only nonsignificant variant exclusively identified by Sanger sequencing was c.1114C>A.
The median TAT of Sanger sequencing was 22 days (range, 14–26 days), while the median TAT of NGS was only 6 days (range, 3–21 days) (Fig. 3). The median difference between Sanger sequencing and NGS TAT was statistically significant (P < 0.001) at 11 days (range, 1–21 days).
Our current study demonstrated the accuracy and short TAT of NGS compared to Sanger sequencing in detecting BRCA1/2 mutations in breast cancer patients. The overall accuracy rate of detecting BRCA1/2 mutations with NGS was 100% with a faster TAT compared to Sanger sequencing. However, some VUS identified by Sanger sequencing were not identically reported by NGS; specifically, many nonsignificant variants that were considered to be no mutation were identified by NGS but not by Sanger sequencing. This discrepancy was likely due to differences in references used for variant nomenclature. Because of the lack of a universally accepted variant reference, a more sophisticated classification of variant nomenclature using existing references is necessary to avoid variant misinterpretation and miscommunication between laboratory investigators and physicians, especially during early phase testing of NGS BRCA1/2 mutation identification.
In this study, some variants with a nonsignificant zygosity possibility not detected by Sanger sequencing were reported in two early NGS cases. To reduce interference, we retested the first 2 NGS samples from these patients, and the nonsignificant zygosity possibility subsequently disappeared. These errors occurred, even though we used a commercially-available BRCA1/2 panel of Ion Torrent sequences, indicating a learning curve exists for achieving the full accuracy of NGS testing.
The sequencing platform, analytical software, and filter settings used can significantly influence the specificity and sensitivity of NGS [811]. Thus, it is important to set appropriate filtering preferences to facilitate more rapid establishment of NGS BRCA1/2 mutation testing. Feliubadaló et al. [11] demonstrated the optimal algorithm with various filter applications of filters in NGS BRCA1/2 testing, and they observed the overall sensitivity and specificity of NGS can differ according to selected filter settings. We have summarized previous studies that evaluated the accuracy of NGS in identifying BRCA1/2 mutations in Table 3 [81112131415]. In general, both sequencing platform and filter settings affected sensitivity and specificity, which, in these studies, ranged from 95% to 100% and 80% to 100%, respectively. In our study, we observed no false-negative or -positive cases when detecting BRCA1/2 mutations by NGS compared to Sanger sequencing, which is consistent with previous study [81112131415]. Therefore, NGS is an alternative diagnostic tool to Sanger sequencing in detecting BRCA1/2 mutations.
Knowing an individual's BRCA1/2 status can critically influence her selection of surgical method related to breast cancer prevention or treatment [5]. Current National Comprehensive Cancer Network guidelines caution women with a known or suspected genetic predisposition to breast cancer against breastconserving surgery [16]. Alternately, contralateral prophylactic mastectomy with ipsilateral mastectomy can be a surgical risk-reduction strategy at the time of definitive surgery, and the rate of performed contralateral prophylactic mastectomy has been increasing in the United States [17]. Therefore, knowing one's genetic susceptibility to breast cancer before surgery and providing corresponding treatment options, such as risk-reduction surgery, in patients with breast cancer or suspicious HBOC syndrome should be achieved as soon as possible. However, it is difficult to obtain this genetic information before surgery in Korea where rapid commercial sequencing services are unavailable. Traditional Sanger sequencing requires a TAT of 4–6 weeks in Korea, and the cost of Sanger sequencing for BRCA1/2 is often between US $1,100–$2,800. Recently, the United States Supreme Court invalidated Myriad Genetics' patent of the BRCA1/2 genes of Myriad in 2013 [18], after which several clinical laboratories in the United States began to offer NGS testing for BRCA1/2 mutations at lower prices than previously available [192021]. This new availability will generate much controversy surrounding BRCA1/2 testing [2021], as the cost for this testing has decreased in other countries other than the United States. In addition, the application of shorter TAT NGS BRCA1/2 testing in high-risk patients with breast cancer compared to Sanger sequencing will change the pattern of clinical practice with respect to surgical option decision-making.
Although we have demonstrated NGS testing is viable in clinical practice, several issues remained to be resolved. Annotation of VUS differs between NGS and Sanger sequencing due to reference database differences if independent laboratories performed the sequencing. This discrepancy makes it difficult for clinicians to provide comprehensive information about VUS for their patients. While increasingly sophisticated methods to address the functional significance of individual VUS are developing [2223], many BRCA1/2 variants remain unclassified [23], and they could be classified differently based which references are used. For example, a previous study reported a reclassification rate of 77% over an 8-year period [24]. Furthermore, the BIC database ceased to be updated by Myriad Genetics after the company stopped contributing their BRCA1/2 results in 2004 [1921]. Therefore, it is not yet possible to report VUS using a universally accepted reference. Using multiple references from international networks to collect and interpret VUS data including clinical, functional, pathological, and in silico analyses may provide a way to share comprehensive BRCA1/2 information [25]. For now, using a single reference for interpreting VUS data should be avoided. Debates regarding the interpretation of VUS are ongoing, and further investigations are needed to establish a universally accepted VUS interpretation method.
National medical systems approving NGS as a screening tool for BRCA1/2 mutation are different among countries. For example, NGS testing for BRCA1/2 mutations is not covered by the Korean national insurance system and is not considered a standard diagnostic tool. Only Sanger sequencing for BRCA1/2 mutations in patients with breast cancer is covered by the national insurance due to a lack of robust validation for BRCA1/2 mutation testing by NGS, although several clinical laboratories in the United States and other Western countries have begun to provide comprehensive BRCA1/2 testing using NGS. However, faster TAT, lower cost, and the technical achievement of managing and exploring NGS data may overcome this hurdle for many countries. Some researchers have highlighted that genomic testing by NGS is very cost-effective and will eventually revolutionize clinical care [26], and a genotype-first approach using NGS is one strategy for managing complex diseases [27]. For these reasons, developing panels for evaluating cancer genes, including BRCA1/2, which are high-penetrance genes and most common in patients and families with HBOC syndrome are critical for positive patient outcomes [2829]. These perspectives suggest that NGS testing for BRCA1/2 and other genes associated with HBOC syndrome will hold a central place in applying novel testing of breast cancer genetics.
Identifying large genomic rearrangements of BRCA1/2 is essential for providing more accurate genomic information to patients. Although commercial BRCA testing detected these rearrangements, neither NGS nor Sanger sequencing detected them in our study. Because conventional Sanger sequencing is not suitable for identifying large genomic rearrangements, additional testing, such as multiplex ligation-dependent probe amplification (MLPA), is necessary in some patients with initially negative BRCA1/2 results as provided by Sanger sequencing [30]. Feliubadaló et al. [11] utilized NGS for identifying large genomic rearrangements, but external validation of this method is still underway in Korea. However, if NGS can detect large genomic rearrangements in BRCA genes without additional methods like MLPA, NGS would be more cost-effective and possibly more accurate than conventional Sanger sequencing.
The shortcoming of this study is its small number of enrolled patients despite its prospective design. A further extension of patient enrollment to ensure data validation is needed. However, this study demonstrates the clear advantage of testing BRCA1/2 using NGS compared to conventional Sanger sequencing. Shorter TAT and high accuracy of NGS may enable clinicians and patients make more timely and informed decisions regarding surgery and neoadjuvant chemotherapy options for preventing or treating breast cancer.
In conclusion, NGS yielded comparably accurate results to Sanger sequencing and in a much shorter time with respect to BRCA1/2 mutation identification. The shorter TAT and higher accuracy of NGS may help clinicians make more timely and informed decisions regarding surgery or neoadjuvant chemotherapy in patients with breast cancer.
Figures and Tables
Fig. 3
Turnaround time (TAT) between next-generation sequencing (NGS) and Sanger sequencing. The dashed and solid lines indicate TAT of Sanger sequencing and NGS, respectively.

ACKNOWLEDGEMENTS
This research was supported by Korea Breast Cancer Foundation and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03934564).
Notes
The protocol of this study was presented at the poster presentation of San Antonio Breast Cancer Symposium 2014, San Antonio, Texas, USA. The abstract of this study was presented at the poster session of Global Breast Cancer Conference 2015 and 4th International Breast Cancer Symposium, Jeju, Korea.
References
1. King MC, Marks JH, Mandell JB; New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003; 302:643–646.
2. Schenberg T, Mitchell G. Prophylactic bilateral salpingectomy as a prevention strategy in women at high-risk of ovarian cancer: a mini-review. Front Oncol. 2014; 4:21.
3. Wevers MR, Aaronson NK, Verhoef S, Bleiker EM, Hahn DE, Kuenen MA, et al. Impact of rapid genetic counselling and testing on the decision to undergo immediate or delayed prophylactic mastectomy in newly diagnosed breast cancer patients: findings from a randomised controlled trial. Br J Cancer. 2014; 110:1081–1087.
4. Evans DG, Lalloo F, Ashcroft L, Shenton A, Clancy T, Baildam AD, et al. Uptake of risk-reducing surgery in unaffected women at high risk of breast and ovarian cancer is risk, age, and time dependent. Cancer Epidemiol Biomarkers Prev. 2009; 18:2318–2324.
5. Schwartz MD, Lerman C, Brogan B, Peshkin BN, Halbert CH, DeMarco T, et al. Impact of BRCA1/BRCA2 counseling and testing on newly diagnosed breast cancer patients. J Clin Oncol. 2004; 22:1823–1829.
6. Zanna I, Rizzolo P, Sera F, Falchetti M, Aretini P, Giannini G, et al. The BRCAPRO 5.0 model is a useful tool in genetic counseling and clinical management of male breast cancer cases. Eur J Hum Genet. 2010; 18:856–858.
7. Metzker ML. Sequencing technologies: the next generation. Nat Rev Genet. 2010; 11:31–46.
8. Chan M, Ji SM, Yeo ZX, Gan L, Yap E, Yap YS, et al. Development of a next-generation sequencing method for BRCA mutation screening: a comparison between a high-throughput and a benchtop platform. J Mol Diagn. 2012; 14:602–612.
9. Han SA, Park SK, Ahn SH, Lee MH, Noh DY, Kim LS, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin Oncol (R Coll Radiol). 2011; 23:434–441.
10. Friedman LS, Gayther SA, Kurosaki T, Gordon D, Noble B, Casey G, et al. Mutation analysis of BRCA1 and BRCA2 in a male breast cancer population. Am J Hum Genet. 1997; 60:313–319.
11. Feliubadalo L, Lopez-Doriga A, Castellsague E, del Valle J, Menendez M, Tornero E, et al. Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet. 2013; 21:864–870.
12. Ozcelik H, Shi X, Chang MC, Tram E, Vlasschaert M, Di Nicola N, et al. Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J Mol Diagn. 2012; 14:467–475.
13. Bosdet IE, Docking TR, Butterfield YS, Mungall AJ, Zeng T, Coope RJ, et al. A clinically validated diagnostic secondgeneration sequencing assay for detection of hereditary BRCA1 and BRCA2 mutations. J Mol Diagn. 2013; 15:796–809.
14. Michils G, Hollants S, Dehaspe L, Van Houdt J, Bidet Y, Uhrhammer N, et al. Molecular analysis of the breast cancer genes BRCA1 and BRCA2 using amplicon-based massive parallel pyrosequencing. J Mol Diagn. 2012; 14:623–630.
15. Hernan I, Borras E, de Sousa Dias M, Gamundi MJ, Mane B, Llort G, et al. Detection of genomic variations in BRCA1 and BRCA2 genes by long-range PCR and next-generation sequencing. J Mol Diagn. 2012; 14:286–293.
16. National Comprehensive Cancer Network. NCCN guidelines Version 3.2014. NCCN guidelines for treatment of cancer by site: breast cancer [Internet]. Fort Wathington (PA): National Comprehensive Cancer Network;c2017. cited 2014 Oct 1. Available from: https://www.nccn.org/professionals/physician_gls/f_guidelines.asp#site.
17. Arrington AK, Jarosek SL, Virnig BA, Habermann EB, Tuttle TM. Patient and surgeon characteristics associated with increased use of contralateral prophylactic mastectomy in patients with breast cancer. Ann Surg Oncol. 2009; 16:2697–2704.
18. Association for Molecular Pathology v. Myriad Genetics, Inc. [Internet]. SCOTUblog;c2017. cited 2017 Apr 3. Available from: http://www.scotusblog.com/case-files/cases/association-for-molecular-pathology-v-myriad-genetics-inc./.
19. Tucker KI. Genetics lab refuses to share data that could save lives; why myriad genetics hoards its information about BRCA mutations [Internet]. New York: The Forward Association Inc;c2017. cited 2014 Sep 27. Available from: http://forward.com/culture/203739/genetics-labrefuses-to-share-data-that-could-save/.
20. So D, Joly Y. Commercial opportunities and ethical pitfalls in personalized medicine: a Myriad of reasons to revisit the Myriad Genetics Saga. Curr Pharmacogenomics Person Med. 2013; 11:98–109.
21. Nelson B. Prometheus bound, but myriad loose ends: amid new legal battles over BRCA tests, technology may resolve what the courts have not. Cancer Cytopathol. 2013; 121:535–536.
22. Domchek SM, Greenberg RA. Breast cancer gene variants: separating the harmful from the harmless. J Clin Invest. 2009; 119:2895–2897.
23. Richter S, Haroun I, Graham TC, Eisen A, Kiss A, Warner E. Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann Oncol. 2013; 24:Suppl 8. viii69–viii74.
24. Murray ML, Cerrato F, Bennett RL, Jarvik GP. Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med. 2011; 13:998–1005.
25. Moghadasi S, Hofland N, Wouts JN, Hogervorst FB, Wijnen JT, Vreeswijk MP, et al. Variants of uncertain significance in BRCA1 and BRCA2 assessment of in silico analysis and a proposal for communication in genetic counselling. J Med Genet. 2013; 50:74–79.
26. Lu JT, Campeau PM, Lee BH. Genotype-phenotype correlation: promiscuity in the era of next-generation sequencing. N Engl J Med. 2014; 371:593–596.
27. Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014; 156:872–877.
28. Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014; 32:2001–2009.
29. Robson M. Multigene panel testing: planning the next generation of research studies in clinical cancer genetics. J Clin Oncol. 2014; 32:1987–1989.
30. Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011; 125:325–349.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download