Abstract
Purpose
The aim of this study is to determine whether levels of circulating free DNA (cfDNA) increase according to cancer progression, whether they are restored after surgical resection, and to evaluate cfDNA in gastric cancer patients as a useful biomarker.
Methods
A case-control study design was used. Thirty gastric cancer patients and 34 healthy subjects were enrolled from two hospitals in South Korea. The plasma cfDNA of patients with gastric cancer were obtained before surgery and 24 hours after surgery, and then analyzed by a quantitative, real-time polymerase chain reaction. Plasma samples were also obtained from the control group.
Results
The mean levels of cfDNA in the healthy control group, patients with early gastric cancer, and with advanced gastric cancer were 79.78 ± 8.12 ng/mL, 106.88 ± 12.40 ng/mL, and 120.23 ± 10.08 ng/mL, respectively (P < 0.01). Sensitivity was 96.67% and specificity was 94.11% when the cutoff value was 90 ng/mL. Variables representing the tumor burden such as tumor size, T stage, TNM stage, and curative resection are also associated with the levels of cfDNA. The levels of cfDNA in the 24-hour-after-surgery group decreased significantly (112.17 ± 13.42 ng/mL vs. 77.93 ± 5.94 ng/mL, P < 0.001) compared to the levels of cfDNA in the preoperation group.
Stomach cancer is the fourth most commonly diagnosed cancer with an average of 1 million patients newly diagnosed annually and was the second leading cause of cancer deaths worldwide in 2008 [1]. Prevalence and mortality are particularly high in East Asia, including South Korea [2]. The 5-year survival rate is more than 90% when cancer is detected as early gastric cancer (EGC), but it decreases to 30%-40% in cases of advanced gastric cancer (AGC) [3]. Thus, early detection and development of gastric cancer biomarkers are important for the treatment and survival of gastric cancer. Biomarkers such as carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 19-9, and CA 72-4 have been investigated for years, but they are not recommended for screening and follow-up of gastric cancer in the National Comprehensive Cancer Network guidelines due to low sensitivity and specificity [4]. This limit of conventional biomarkers requires development of novel cancer biomarkers.
As a cancer cell develops, circulating free DNA (cfDNA) is released into the blood stream with various physiologic events such as micrometastasis, necroptosis, apoptosis, and secretion [5]. CfDNA from the physiologic apoptosis of cells can be detected in healthy subjects [6] and is slightly prone to be increased in cases of trauma, sepsis, and other diseases such as systemic-lupus syndrome, pulmonary embolism, and myocardial infarction [7]. However, cfDNA in the plasma of cancer patients is 2-3 times higher than in normal healthy groups in various cancer studies [8]. Many studies suggest that circulating tumor cells or cfDNA can be alternative biomarkers for early detection of cancers, and predict prognosis and efficacy of therapies [8]. This cfDNA is also increased in benign preneoplastic tumors. Previous studies demonstrate that cfDNA is proportionally increased in benign colon polyps and colon cancers individually compared to normal groups [9]. Decreased cfDNA is mainly attributed to reduced tumor size by an operation and also rapid clearance by plasma nuclease such as DNAseI [10]. Previous studies demonstrated a half-life of cfDNA in plasma that ranges from minutes to hours. CfDNA can be a useful biomarker for the prediction of prognosis or cancer relapse. For example, one study demonstrated that the 2-year survival rate was 48% in cfDNA-detected groups and 100% in cfDNA-nondetected groups among colon cancer patients [11].
As many research outcomes point out cfDNA as a significant marker, cfDNA could be a good marker for early detection and follow-up treatment in gastric cancer. The purpose of this study was to determine the implication of cfDNA as a useful biomarker to detect EGC, to predict tumor burden and to detect residual tumor after surgery.
A cross-sectional case-and-control study design was applied to two Korean hospitals, located in a metropolitan area, between October 2012 and March 2013.
Thirty gastric cancer patients who underwent a gastrectomy with a curative intent in Seoul Medical Center, and 34 age-matched healthy controls who visited for regular health check-ups in MizMedi Hospital, were recruited. A healthy control was defined as a subject who was not diagnosed of any cancerous condition in the past and currently does not have a serious illness, such as severe infection, sepsis, or trauma.
The case group is defined as patients who had been diagnosed with gastric cancer, which was pathologically confirmed by an endoscopic biopsy. Patients underwent either a laparoscopy or open surgery. Patients who underwent a palliative resection or who had prior chemotherapy, distant metastasis and double-primary cancer, were excluded. Data obtained for each patient included age, sex, body mass index (BMI), tumor marker (CEA, CA 19-9), tumor size, histologic type, T stage, N stage, gastric cancer stage classified according to the seventh edition of the American Joint Committee on Cancer staging criteria [12], and the preoperative and post-24-hour operative serum cfDNA level. In the control group, healthy people who underwent an endoscopy for cancer screening were selected in order to obtain data.
Many procedures, such as phenol-chloroform extraction, salting-out, magnetic beads, and triton/heat/phenol protocols [13] have been used to isolate cfDNA. To successfully isolate cfDNA from plasma, we empirically found that the efficacy of the extraction procedures is the key issue. We compared the efficacy of several commercial cfDNA-isolation kits based on column-based systems such as QIAamp DNA Micro kit (Qiagen, Valencia, CA, USA), Nucleospin Plasma XS (Macherey-Nagel GmbH & Co. KG, Düren, Germany), and G-spin Total DNA Extraction kit (Intron biotechnology, Seongnam, Korea). Comparing plasma DNA yield and extent of DNA concentration variation from sample to sample, the QIAamp DNA Micro kit showed least variety in the amount of cell-free DNA among the 3 kits (data not shown). We reached the conclusion that the QIAamp DNA Micro kit provided robust and reliable cfDNA isolation. We routinely used 1.5-mL plasma as starting material, and the DNA was extracted according to the protocols instructed by the supplier. All blood samples were centrifuged once for 10 minutes at 4,000×g. From the withdrawn plasma, 200 µL of plasma were dispensed to 7 microcentrifuge tubes per sample. Twenty-µL proteinase K and 4 µL of an RNase A stock solution were added to each tube. Then, 200 µL of Buffer AL was added to the sample and mixed thoroughly to yield a homogeneous solution. The tube was incubated at 56℃ for 10 minutes. Absolute ethanol (200 µL) was added to each reaction tube and then mixed by pulse-vortexing. For each sample, a 7 microcentrifuge mixture was then applied to the same QIAamp mini spin column to increase the recovery yield and centrifuged at 9,000 rpm for 1 minute. Then the filtrate was discarded. Afterwards, 500 µL of Buffer AW1 was carefully added to the spin column and centrifuged at 9,000 rpm for 1 minute. Here again, the filtrate was discarded. Then 500 µL Buffer AW2 was added to the spin column and centrifuged at 13,000 rpm for 1 minute. The QIAamp mini spin column was applied to a new collection tube and centrifuged at 13,000 rpm for 1 minute again. Finally, the spin column was placed in a clean 1.5-mL microcentrifuge tube and 50-µL Buffer AE was added. After incubation at room temperature for 5 minutes, it was then centrifuged at 13,000 rpm for 1 minute to elute the DNA.
Descriptive and comparative analyses were done. Fisher exact test, Wilcoxon rank sum test, and the Kruscal-Wallis test were used to compare demographics and clinical characteristics. Post hoc multiple comparisons were calculated using Tukey method. A receiver-operating characteristic (ROC) curve was generated to assess the cfDNA level as a diagnostic biomarker. A cutoff point was chosen, and then sensitivity, false-positive rate, and 95% confidence intervals were calculated. A linear-regression model was used to adjust the covariants (including age and sex) affecting cfDNA. All tests were analyzed by IBM SPSS ver. 20.0 (IBM Co., Armonk, NY, USA).
This study was conducted prospectively after receiving permission from the local Institutional Review Board (No. 2012-056). All patients participated voluntarily with written informed consents.
The mean age was 66.72 ± 13.16 years in 30 gastric cancer patients, and 63.79 ± 6.76 years in 34 age-matched healthy subjects (P = 0.256). Aging cfDNA increases in gastric cancer patients (P < 0.01), but age and cfDNA are not correlated in the healthy control group (P = 0.969). CfDNA in females was higher than males in gastric cancer patients (P = 0.01), but not in the healthy control group (P = 0.598). CfDNA in nonsmokers was higher than smokers (P = 0.033) in gastric cancer patients, but not in the healthy control group (P = 0.375). Drinking alcohol, BMI, and Helicobacter pylori infection are not associated with cfDNA both in gastric cancer patients and the healthy control group (Table 1).
CfDNA was proportionally increased between healthy subjects, EGC patients, and AGC patients (79.78 ± 8.12 ng/mL, 106.88 ± 12.40 ng/mL, and 120.23 ± 10.08 ng/mL, P < 0.001) (Fig. 1, Table 2). Fig. 2 shows the ROC curves of cfDNA between the cancer patients and healthy controls. The area under the curve is 0.991. As the cutoff value of cfDNA is defined to 90 ng/mL, sensitivity, specificity, positive-prediction value, and negative-prediction value are 96.67%, 94.11%, 93.54%, and 96.96%, respectively.
Table 2 shows clinical data that represent tumor burden. cfDNA is higher in the AGC group than the EGC group, and this is significant after adjustment by age, sex, and smoking (P = 0.004 and P = 0.035, respectively). CfDNA in the group with tumor sizes 5-9.9 cm is higher than in the group with tumor sizes <4.9 cm (P = 0.280, Tukey multiple-comparison test). Differentiation is not associated with cfDNA. The association between cfDNA and T stage is less significant (P = 0.065) but becomes significant after adjustment by age, sex and smoking (P = 0.037). N stage is not associated with cfDNA. CfDNA in high-TNM stage is higher than in low-TNM stage and is significant after adjustment for age, sex and smoking (P = 0.048). Variable of curability is significantly associated with cfDNA when unadjusted and adjusted, respectively (P < 0.001 and P = 0.016).
CfDNA is not associated with the tumor marker of CEA. All CA 19-9 in patients with gastric cancer were within normal ranges, and correlation between CA 19-9 and cfDNA is not significant (data are not shown). CEA and CA 19-9 are not associated with variables that represent the tumor burden such as cancer progression (EGC vs. AGC) and TNM stage (data are not shown).
Compared to the mean levels of cfDNA in the preoperation group, the mean levels of cfDNA in the 24-hour-after-surgery group decreased significantly (112.17 ± 13.42 ng/mL vs. 77.93 ± 5.94 ng/mL, P < 0.001) (Fig. 3).
There is an increased incidence of stomach cancer cases in Korea, indicating a need for an applicable biomarker for better screening and management. According to the Korean National Cancer Incidence Database, in 2010, 202,053 incidents of cancer cases and 72,046 deaths were identified in Korea. The most prevalent cancer was thyroid cancer (36,021 cases), and the next most prevalent cancer was stomach cancer (total 30,092 cases: 2,017 males and 9,913 females). A total of 10,032 patients died due to stomach cancer in 2010. The crude rate of cancer incidence was 259.9 per 100,000 in the general Korean population. The age-standardized incidence rate of gastric cancer per 100,000 populations was 41.8 (males 62.3, females 24.9) in 2010 in Korea [14]. Also, Park et al. [15] reported that the incidence of second primary cancer (SPC) in male cancer survivors was 603.2 per 100,000 person-years, which was about 2.3 times compared to the incidence of the general population.
Thus, the incidence of gastric cancer and the SPC risk of cancer survivors in South Korea are much higher than in other countries. Although health screening for early diagnosis and follow-up tools are high, the use of conventional tumor markers (CEA, CA 19-9, CA 72-4, etc.) has many limits due to low sensitivity and specificity. So, more sensitive and specific biomarkers should be used for stomach cancer detection in Korea.
In this pilot study, we evaluated the efficacy of cfDNA as a biomarker that discriminates cancer patients from healthy subjects. The main finding of this study was that the plasma level of cfDNA is able to differentiate tumor burden such as tumor size, depth of invasion, and tumor stage. In our study, some demographic characteristics of patients with gastric cancer, such as age, sex and smoking, are associated with plasma-mean cfDNA levels. An unexpected result is that the levels of cfDNA are higher in the nonsmoking groups than the smoking groups. However these variables are not different in the healthy control group (Table 1). This may be due to the fact that the cancer type in the particular variables such as aged, female, non-smoker is more aggressive than others in this small sampled study.
Many studies have demonstrated that cfDNA levels have discriminatory power to differentiate cancer patients from healthy controls. As a diagnostic tool, sensitivity and specificity of cfDNA between cancer patients and healthy controls varies. Kamat et al. [16] demonstrated that a cutoff value of 4,500 GE/mL of cfDNA yielded a sensitivity of 87% and a specificity of 87% among 164 women with invasive epithelial ovarian carcinoma, 49 with benign ovarian neoplasms, and 75 age-matched controls. Another study showed cfDNA from plasma in metastatic colorectal cancer patients yields a specificity of 97% and a sensitivity of 31% [17]. Our study showed a much higher diagnostic value both in sensitivity and specificity compared to previous studies.
Furthermore, cfDNA can be a possible biomarker to differentiate the invasiveness of tumors. Increased serum cfDNA levels in relationship to tumor size are predictive of distant metastasis of esophageal squamous cell carcinoma [18]. Agostini et al. [19] demonstrated that cfDNA in breast cancer patients is associated with lymph-node involvement, but not with tumor stage and vascular invasion. The level of cfDNA was associated with malignant tumor size, lymph-node involvement, stage, and grade, as well as Her2/neu and Topoisomerase IIα expression, in breast cancer patients [20]. This study also demonstrated that the levels of cfDNA in plasma are proportionally associated with parameters representing the invasiveness of gastric tumors such as clinical cancer type (EGC vs. AGC), tumor size, and tumor stage, but not with histological differentiation and lymph-node involvement. In contrast to the cfDNA in this study, CEA and CA 19-9 are not associated with tumor stage and histological tumor type (data not shown). Although tumor markers such as CEA and CA 19-9 have been used in AGC, most of them are not recommended for screening for early detection or posttreatment surveillance [21]. Clinically, a biomarker of cfDNA can be applied to search distant metastasis and microinvasion of tumors, and it eventually helps to select treatment choices according to molecular diagnosis [22].
Our study also demonstrates that levels of cfDNA are significantly decreased 24 hours after a surgical resection. Many studies have shown that cfDNA is a good biomarker for monitoring treatment response and detecting recurrence after treatment. One study evaluated cfDNA at one week after a surgical resection of colorectal cancer and followed up in monthly intervals [23]. Levels of cfDNA also reflect the response after chemotherapy in many studies. After preoperative chemotherapy, the index of cfDNA integrity decreased in patients with rectal cancer that responded to chemotherapy compared to subjects that did not respond to therapy [24]. High cfDNA in plasma is associated with metastasis of tumors and poor survival outcomes in patients with advanced lung cancer [25].
There is no agreement among the methods in which the extraction protocol is consistent, reproducible, and reliable. This may be due to considerable variation among studies including specimen type (plasma or serum), duration from collection to extraction, and cfDNA-isolation methodology. By avoiding red blood cell lysis (hemolysis) and concomitant contamination with DNA derived from nucleated cells, we have specifically isolated the cfDNA from the plasma. Meanwhile, previous studies revealed that serum is a less-suitable specimen type because it becomes readily contaminated with DNA from white blood cells when blood coagulation is evoked [26].
This study has some limitations. First, the limit of sample numbers in each category of tumor stages seems to be attributed to the statistical nonsignificance. Second, data of healthy controls does not include a tumor marker-CEA, CA 19-9-and thus we couldn't directly compare such conventional biomarkers with cfDNA. Third, in order to evaluate the relevance of cfDNA to survival, we need to follow up these gastric cancer patients further. Careful randomized prospective trials and comparison with existing established factors will be required to develop cfDNA as a useful cancer biomarker for cancer screening, treatment monitoring, and prognostic markers. Further, qualitative DNA assays, including genetic or epigenetic alteration such as DNA methylation of tumor-suppressor genes, need to be investigated in surrogating tissue biopsy.
In conclusion, cfDNA can be a valuable biomarker for early cancer detection and evaluating tumor burdens. Also, with further studies, cfDNA could be a marker for estimating completeness of surgical resection in patients with gastric cancer.
Figures and Tables
 | Fig. 1Comparison of circulating free DNA among healthy subjects (white), early gastric cancer (EGC) groups (gray), and advanced gastric cancer (AGC, black) groups. |
 | Fig. 2Receiver operating characteristic (ROC) curve of circulating free DNA between cancer patients and healthy controls. AUC, area under the curve. |
 | Fig. 3Comparison of mean circulating free DNA levels between pre- and postoperation in patients with gastric cancer. |
Table 1
Mean levels of cfDNA according to clinical charateristics in gastric cancer and healthy control group

Table 2
Correlation between cfDNA and clinico-pathological data, which represents the invasiveness of gastric cancer
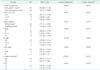
cfDNA, circulating free DNA; SD, standard deviation; CEA, carcinoembryonic antigen.
a)Generalized linear-regression model; variables of age, sex, and smoking were adjusted. b)Curative resection means R0 resection (no residual tumor), R1 (microscopic residual tumor), nd R2 resections (macroscopic residual tumor) are included in the noncurative resection groups. c)Total number of cases with preoperative CEA level checked were 27 cases.
References
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008: Cancer Incidence and Mortality Worldwide [Internet]. Lyon: International Agency for Research on Cancer;c2010. cited 2013 Aug 30. Available from: http://www.iarc.fr/en/media-centre/iarcnews/2010/globocan2008.php.
2. Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, et al. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009; 125:666–673.
3. Wang J, Yu JC, Kang WM, Ma ZQ. Treatment strategy for early gastric cancer. Surg Oncol. 2012; 21:119–123.
4. Strong VE, D'Amico TA, Kleinberg L, Ajani J. Impact of the 7th Edition AJCC staging classification on the NCCN clinical practice guidelines in oncology for gastric and esophageal cancers. J Natl Compr Canc Netw. 2013; 11:60–66.
5. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–437.
6. Anker P, Stroun M. Circulating DNA in plasma or serum. Medicina (B Aires). 2000; 60(5 Pt 2):699–702.
7. Cui M, Fan M, Jing R, Wang H, Qin J, Sheng H, et al. Cell-Free circulating DNA: a new biomarker for the acute coronary syndrome. Cardiology. 2013; 124:76–84.
8. De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol. 2011; 7:1385–1397.
9. Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer. 2011; 105:239–245.
10. Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007; 635:105–117.
11. Lecomte T, Berger A, Zinzindohoue F, Micard S, Landi B, Blons H, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002; 100:542–548.
12. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC cancer staging manual. 7th ed. New York: Springer;2010.
13. Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009; 404:100–104.
14. Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG, Lee JS. Cancer statistics in Korea: incidence, mortality, sur vival and prevalence in 2010. Cancer Res Treat. 2013; 45:1–14.
15. Park SM, Lim MK, Jung KW, Shin SA, Yoo KY, Yun YH, et al. Prediagnosis smoking, obesity, insulin resistance, and second primary cancer risk in male cancer survivors: National Health Insurance Corporation Study. J Clin Oncol. 2007; 25:4835–4843.
16. Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010; 116:1918–1925.
17. Morgan SR, Whiteley J, Donald E, Smith J, Eisenberg MT, Kallam E, et al. Comparison of KRAS mutation assessment in tumor DNA and circulating free DNA in plasma and serum samples. Clin Med Insights Pathol. 2012; 5:15–22.
18. Tomochika S, Iizuka N, Watanabe Y, Tsutsui M, Takeda S, Yoshino S, et al. Increased serum cell-free DNA levels in relation to inflammation are predictive of distant metastasis of esophageal squamous cell carcinoma. Exp Ther Med. 2010; 1:89–92.
19. Agostini M, Enzo MV, Bedin C, Belardinelli V, Goldin E, Del Bianco P, et al. Circulating cell-free DNA: a promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012; 11:89–98.
20. Hashad D, Sorour A, Ghazal A, Talaat I. Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal. 2012; 26:467–472.
21. Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014; 17:26–33.
22. Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010; 17:R245–R262.
23. Levy M, Benesova L, Lipska L, Belsanova B, Minarikova P, Veprekova G, et al. Utility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patients. Anticancer Res. 2012; 32:1621–1626.
24. Agostini M, Pucciarelli S, Enzo MV, Del Bianco P, Briarava M, Bedin C, et al. Circulating cell-free DNA: a promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann Surg Oncol. 2011; 18:2461–2468.
25. Lee YJ, Yoon KA, Han JY, Kim HT, Yun T, Lee GK, et al. Circulating cell-free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first-line therapy. Clin Cancer Res. 2011; 17:5179–5187.
26. Umetani N, Hiramatsu S, Hoon DS. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci. 2006; 1075:299–307.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download