Abstract
Purpose
Recently, two alternatively spliced survivin variants, survivin-ΔEx3 and survivin-2B, were identified in a single copy of the survivin gene. It has been reported that the expressions of survivin splice variants significantly correlates with the clinical results in many types of human carcinoma. We investigated the transcription levels of survivin and its splice variants in human colorectal carcinomas, and analyzed correlations between survivin expression levels and clinicopathologic features.
Methods
We used Western blot and real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) to analyze the protein and mRNA expression levels of survivin variants in 51 colorectal carcinomas. The quantitative RT-PCR was performed using primer pairs specific for survivin and each of its splice variants, then normalized for the gene that encodes glyceraldehydes-3-phosphate dehydrogenase.
Results
In Western blotting, the protein levels of survivin were higher in the tumor tissue than in normal tissue. The expression of survivin, survivin-2B and survivin-ΔEx3 mRNA was present in 96%, 64.7%, and 82.4% of the samples, respectively. When the pathologic parameters were compared, colorectal cancers of advanced pT stages showed significant decrease in survivin-2B mRNA expression by the quantitative RT-PCR (P < 0.001).
Genome-wide analysis of alternative splicing has indicated that 35 to 59% of human genes have alternatively spliced forms, and that 70 to 88% of alternative splices generates a multitude of variants that have overlapping but distinct functions [1]. Alternatively spliced forms of key proteins in cancer, such as TP53, MDM2, and c-MYC, have been shown to play roles in oncogenesis [2-4].
Survivin, which is a novel member of the inhibitor of apoptotic protein (IAP) family, is markedly over-expressed in most types of human carcinomas. It has been investigated with regard to its correlation with clinical results and its potential as a target for anticancer therapy [5].
Two alternatively spliced survivin variants, survivin-ΔEx3 and survivin-2B, were recently identified in a single copy of the survivin gene [6]. Currently, there is no strong evidence to suggest that survivin splice variants are involved in tumorigenesis. However, there have been reports that the expression of survivin splice variants significantly correlates with the clinical results in many types of human carcinoma, including neuroblastoma, renal cell carcinoma, gastric cancer, brain tumor, leukemia, and breast cancer. Many investigators have suggested that survivin-ΔEx3 and survivin-2B play opposing roles in tumor progression and/or tumorigenesis [7-12].
The over-expression of survivin in colorectal carcinoma has been identified as an independent prognostic factor [13,14]. However, few reports had addressed on the correlation between survivin splice variants and the clinical results of colorectal cancer [15].
Therefore we investigated the expression levels of survivin and its splice variants in human colorectal carcinomas and assessed the relationship between the expression of survivin splice variants and the clinicopathologic parameters.
The present study included 51 primary colorectal carcinoma patients who were admitted between January 2003 and December 2004. These patients had not received chemotherapy or radiation therapy before undergoing tumor resection. There were 35 males and 16 females, and the median age of the patients was 65 years (range, 32 to 92 years). According to the American Joint Committee on Cancer classification, the numbers of patients with stage I, II, and III cancer were 19, 9, and 23, respectively.
We obtained paired colorectal cancerous and adjacent normal mucosa from each patient at the time of surgery. The samples were immediately frozen in liquid nitrogen and stored at -80℃ until use.
Fresh frozen tissues were evenly ground with a homogenizer, and cell lysis was performed with the Pro-PREP Protein Extraction solution (Intron Biotechnology, Seongnam, Korea) in a cold room at 4℃. Protein extracts were obtained by centrifugation at 14,000 rpm for 5 minutes at 4℃. The protein content was measured by the Bradford assay method, and the samples were heated for 8 minutes for denaturation. Next, 40 mg of protein was electrophoresed in a 14% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose membrane. The blot was incubated in blocking buffer (4% nonfat milk, 20 mM Tris [pH 7.5], 500 mM NaCl, 0.1% Tween-20) for 1 hour, and was incubated with affinity-purified rabbit anti-human survivin antibody (1:1,000 dilution; R&D Systems Inc., Minneapolis, MN, USA) overnight at 4℃. After three washes for 10 minutes each, anti-rabbit Ig horseradish peroxidase-conjugated whole antibody from donkey (1:2,000; Amersham Biosciences, Uppsala, Sweden) was added, and was incubated for 1 hour. The blot was then washed three times and the bands were detected with the ECL Plus kit (Amersham Biosciences).
The RNAs from the colorectal cancer tissues, which were stored at -80℃, were extracted using totally RNA (Ambion, Austin, TX, USA) and purified with the RNeasy kit (Qiagen, Hilden, Germany), to eliminated DNA contamination. The integrity of the extracted RNA was analyzed by agarose electrophoresis and confirmed by intact 18S/28S rRNA bands. The RNA was reverse-transcribed together with oligo (dT) primer using the Power cDNA Synthesis kit (Intron Biotechnology). First, 1 µg of RNA was incubated in 20 µL of a solution that contained 1 µL RNase inhibitor, 4 µL of 5 × reverse transcriptase (RT) buffer, 2 µL dNTP, 2 µL dithiothreitol, 0.5 µL avian myeloblastosis virus RT enzyme at 42℃ for 60 minutes. The reaction was ended after incubation at 70℃ for 5 minutes.
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) was performed in triplicate in 384-well plates; each 20-µL reaction consisted of 10 µL of SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), 0.8 µL of 10 pM forward and reverse primers for the survivin gene, its splice variants or the glyceraldehydes-3-phosphate dehydrogenase (GAPDH) gene. The PCR conditions were 50℃ for 2 minutes and 95℃ for 10 minutes, followed by 40 cycles of 95℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 30 seconds. The sequence of the common forward primer for the survivin variants was 5'-GACCACCGCATCTCTACATTC-3'. To distinguish the three splice variants of survivin, the following reverse primers were used: for survivin, 5'-TGCTTTTTATGTTCCTCTATGGG-3'; for survivin-2B, 5'-AAGTGCTGGTATTACAGGCGT-3'; and for survivin-ΔEx3, 5'-ATTGTTGGTTTCCTTTGCATG-3'. The primers for GAPDH were: forward, 5'-TTGGTATCGTGGAAGGACTCA-3'; and reverse, 5'-TGTCATCATATTTGGCAGGTTT-3'. Each of the 384-well quantitative RT-PCR plates included serial dilutions (1, 0.5, and 0.25) of cDNA, which were used to generate relative standard curves for the survivin variant genes and GAPDH. The GAPDH copy numbers (external standard) were used to normalize the copy numbers of the different survivin variants, by calculating the ratios of the relative mRNA levels.
The data of relative gene expression was analysed using the 2-ΔΔCt method, where ΔΔCt = (Ct,Target - Ct,GAPDH)Time× - (Ct,Target - Ct,GAPDH)Time 0. The Time x is any time point and Time 0 represents the 1X expression of the target gene normalized to GAPDH. The real-time RT-PCR analysis was performed on the Prism 7900 Sequence Detection System (Applied Biosystems).
The relative amounts of survivin variant mRNAs were determined by dividing the amount of the survivin variant mRNAs by the amount of GAPDH mRNA for each sample. We examined the potential correlation between the transcription expression levels of the survivin variants and clinicopathologic factors.
In the Western blots of the colorectal cancer tissues, single discrete band of approximately 18.1 kDa was observed, indicating the presence of the survivin protein. The tumor tissues express survivin 10 to 15 times more than normal ones (Fig. 1). The specificities of the amplification products were confirmed by agarose gel electrophoresis, in which distinct bands of the calculated sizes were detected for all PCR products at the end-point (Fig. 2). Among the 51 tumor samples, survivin mRNA was the most dominant transcript and was present in 96% (49/51) of the samples; 64.7% (33/51) of the samples demonstrated survivin-2B expression, and 82.4% (41/51) of the samples showed survivin-ΔEx3 expression.
The expression of survivin-2B decreased along with the invasive depth of the tumor with statistical significance (P < 0.001), while the expression levels of survivin and survivin-ΔEx3 did not show any relationship with any of the investigated clinicopathologic factors (Table 1, Fig. 3).
To ensure that the depth-dependent decrease of survivin-2B expression was not the result of a depth-dependent increase of the GAPDH transcript used as the external standard, we also calculated the ratios of the mRNA levels between survivin-ΔEx3 and survivin-2B on the one hand, and survivin as an internal standard on the other hand. Since all of the survivin variants are derived from a common hnRNA precursor pool, these ratios are independent of any potential bias imposed by possible variations of the GAPDH expression levels. We did not find any significant change in the relative mRNA levels between survivin and its variants, which supports the observed depth-dependent decrease in survivin-2B expression (Table 1, Fig. 4).
Colorectal cancer is one of the leading causes of death worldwide, and many researches have been performed on the development and progression of this tumor. The development of colorectal cancer proceeds through a series of genetic changes that involve the activation of oncogenes and the loss of tumor suppressor genes. During this process, a disturbance of the balances between cell proliferation and apoptosis may underlie neoplastic development [16]. Cell death is caused by necrosis and apoptosis, with apoptosis playing an important role in homeostatic maintenance in mature human organs by removing senescent and unneeded cells [17]. Unfortunately, cancerous cells may continue to grow by inhibiting the apoptosis induced by natural defense products, such as bcl-2 [18], heat-shock protein [19], and members of the IAP family [20]. In addition, these antiapoptotic products may facilitate the accumulation of transforming mutations and promote resistance to anticancer chemotherapy [21,22].
Among the antiapoptotic products, the IAP family has been recently discovered. IAP family members directly inhibit activated caspase 3 and caspase 7, and inhibit the activation of pro-caspase 9. There are several lines of evidence that they also indirectly inhibit caspase 1 and caspase 2 as well [21,22]. The IAP family has been shown to protect cells from various types of apoptosis inducing agents and to block apoptosis generated by various stimuli, such as various anticancer drugs, irradiation, and ultraviolet irradiation; thus, the IAP family plays a key role in the formation of cancer [19].
The locus of survivin, which is a recently discovered member of the IAP family, is at 17q25. Survivin consists of 142 amino acids and has a molecular mass of 16.5 kDa [23]. Unlike other members of the IAP family, survivin shows high-level expression during the fetal period and negligible expression in normally differentiated tissues after birth. However, studies have shown that the expression of survivin increases in cancer cells [20]. Discoveries of the expression of survivin in tissues of various cancers, such as lung, colon, stomach, breast, prostate, and pancreatic cancers, as well as neuroblastomas and highly differentiated lymphomas have brought survivin to the forefront of research efforts [5].
Survivin mRNA has been detected in 67.3% of colon cancer tissues, and the expression of survivin has been found to be significantly higher in colon cancer tissues [14] compared to the normal tissue. The human survivin genes have four dominant (1, 2, 3, and 4) and two hidden (2B and 3B) exons. Their own pre-mRNA creates four mRNAs with different conjugations of splices, which are designated as the survivin, survivin-2B, survivin-ΔEx3, and survivin-3B proteins [6]. Survivin-ΔEx3, which lacks exon 3, and survivin-2B, which has intron 2 as an additional cryptic exon, has been studied in detail [6,24]. In the colon mucosa, survivin is preferentially expressed in the lower portion of normal colon crypts where adult stem cells and other proliferative cells reside and its expression decreases from crypt bottom to top as the cells progress to differentiate [25]. The survivin intensively expressed in many fetal tissues in contrast with downregulation in most nonproliferating adult tissues. However, reexpression of survivin was found in the most common human cancer types, which might promote both tumor progression and resistance to treatment with anticancer drugs and irradiation. The biochemical a mechanism by which survivin suppresses apoptosis remains under investigation.
In this study, the survivin-2B was decreased in advanced pT3 stages in contrast with the relative increase in early stages (pT1 + 2). The downregulation of survivin-2B might weaken its functional antagonism, and moreover, could permit the generation of more survivin or survivin-ΔEx3 because three survivin variants come from the same pool of an hnRNA precursor. Thus, the decrease in survivin-2B may result in development and invasion of the colorectal carcinoma. Mahotka et al. [8] and Meng et al. [9] also reported that survivin-2B decreased according to the higher clinical stages and invasive depth in renal cell carcinoma and gastric carcinoma, respectively.
Islam et al. [7] have reported that high level of survivin mRNA expression were significantly associated with advanced stages of neuroblastomas; survivin-2B expression was predominant in some favorable neuroblastoma, while it was low and ubiquitous in most normal and malignant tissues. Krieg et al. [26] have reported that survivin-2B is significantly decreased in the later stages of stomach cancer. Meng et al. [9] have reported that survivin-2B is significantly related to the initial stage, differentiation, and infiltration of the lesion among stomach cancer patients, while survivin-ΔEx3 is significantly correlated with apoptosis. Nakagawa et al. [11] have reported that survivin-ΔEx3 is more conspicuous than survivin-2B in the bone marrow tissues of acute and chronic lymphatic leukemia patients. In summary, researchers have pointed out that survivin-ΔEx3 show a clear antiapoptotic function, while survivin-2B greatly decreases the antiapoptotic function of survivin. Therefore, survivin-ΔEx3 and survivin-2B play important opposite roles in the generation and progression of tumors.
Recently, Suga et al. [15] have reported the correlation of the stages in patients of colorectal cancer with clinicopathologic factors. According to this study, GAPDH-normalized mRNA of survivin and its splicing variants have no correlated with the stage, survivin-2B/survivin has the only correlation. In this study, we also could not find the correlation with stages. However we subdivided to T and N stage and analyzed the correlation respectively. Consequently, we found that the level of GAPDH-normalized survivin-2B mRNA decreases significantly as the depth of the lesion increases.
In conclusion, survivin was expressed to a greater extent in colon cancer tissues than in normal tissues. However, when survivin and its variants were separated and the quantities of mRNA were measured, only survivin-2B showed a significant difference in expression, which was dependent upon the depth of the lesion and appeared to correlate with the progression of colon cancer.
This study presents that the expression of survivin-2B decreases according to the depth of invasion in the colorectal carcinoma, although those of survivin and survivin-ΔEx3 have not related with the tumor progression and stage. Determining the expression pattern of survivin variants in each sample might have a practical usefulness in better classifying patients whom would be subjected to therapies based on targeting survivin mRNA or protein. Further studies for elucidating the exact role of survivin variants in tumorigenesis of colorectal tissues along with their roles related to recurrence, progression, and patient survival are needed.
Figures and Tables
Fig. 1
Western blotting for survivin in colon cancer tissues. Normal colon (N) and colon cancer (T) tissues were analyzed in a 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel. Single and distinct bands of approximately 18.1 kDa are evident, indicating the presence of the survivin protein.
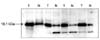
Fig. 2
Specificities of the amplification products detected by agarose gel electrophoresis, with distinct bands of the calculated sizes for all polymerase chain reaction (PCR) products at the end-point of 50 PCR cycles. GAPDH, glyceraldehydes-3-phosphate dehydrogenase.
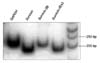
Fig. 3
GAPDH-normalized mRNA levels of survivin and its variants in colorectal cancers in relation to the different variables. A significant decrease in the expression of survivin-2B is noted in advanced T stage (pT3) patients. Top line representslargest value, bottom line represents smallest value, the upper line of the box represents upper quartile, the lower line of the box represents lower quartile, and inner line of the box represents median value.

Fig. 4
Ratios of the relative mRNA levels between survivin and its variants in colorectal cancer. No significant change is evident in relation to T stage. Top line represents largest value, bottom line represents smallest value, the upper line of the box represents upper quartile, the lower line of the box represents lower quartile, and inner line of the box represents median value.

References
1. Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002. 30:13–19.
2. Ng CC, Koyama K, Okamura S, Kondoh H, Takei Y, Nakamura Y. Isolation and characterization of a novel TP53-inducible gene, TP53TG3. Genes Chromosomes Cancer. 1999. 26:329–335.
3. Weng MW, Lai JC, Hsu CP, Yu KY, Chen CY, Lin TS, et al. Alternative splicing of MDM2 mRNA in lung carcinomas and lung cell lines. Environ Mol Mutagen. 2005. 46:1–11.
4. Yukitake H, Furusawa M, Taira T, Iguchi-Ariga SM, Ariga H. AAT-1, a novel testis-specific AMY-1-binding protein, forms a quaternary complex with AMY-1, A-kinase anchor protein 84, and a regulatory subunit of cAMP-dependent protein kinase and is phosphorylated by its kinase. J Biol Chem. 2002. 277:45480–45492.
5. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997. 3:917–921.
6. Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005. 92:212–216.
7. Islam A, Kageyama H, Hashizume K, Kaneko Y, Nakagawara A. Role of survivin, whose gene is mapped to 17q25, in human neuroblastoma and identification of a novel dominant-negative isoform, survivin-beta/2B. Med Pediatr Oncol. 2000. 35:550–553.
8. Mahotka C, Krieg T, Krieg A, Wenzel M, Suschek CV, Heydthausen M, et al. Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer. 2002. 100:30–36.
9. Meng H, Lu C, Mabuchi H, Tanigawa N. Prognostic significance and different properties of survivin splicing variants in gastric cancer. Cancer Lett. 2004. 216:147–155.
10. Yamada Y, Kuroiwa T, Nakagawa T, Kajimoto Y, Dohi T, Azuma H, et al. Transcriptional expression of survivin and its splice variants in brain tumors in humans. J Neurosurg. 2003. 99:738–745.
11. Nakagawa Y, Yamaguchi S, Hasegawa M, Nemoto T, Inoue M, Suzuki K, et al. Differential expression of survivin in bone marrow cells from patients with acute lymphocytic leukemia and chronic lymphocytic leukemia. Leuk Res. 2004. 28:487–494.
12. O'Driscoll L, Linehan R, M Kennedy S, Cronin D, Purcell R, Glynn S, et al. Lack of prognostic significance of survivin, survivin-deltaEx3, survivin-2B, galectin-3, bag-1, bax-alpha and MRP-1 mRNAs in breast cancer. Cancer Lett. 2003. 201:225–236.
13. Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998. 58:5071–5074.
14. Chen WC, Liu Q, Fu JX, Kang SY. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol. 2004. 10:2886–2889.
15. Suga K, Yamamoto T, Yamada Y, Miyatake S, Nakagawa T, Tanigawa N. Correlation between transcriptional expression of survivin isoforms and clinicopathological findings in human colorectal carcinomas. Oncol Rep. 2005. 13:891–897.
16. Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995. 267:1456–1462.
17. Rudin CM, Thompson CB. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med. 1997. 48:267–281.
18. Yang E, Korsmeyer SJ. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996. 88:386–401.
19. Jäättelä M. Escaping cell death: survival proteins in cancer. Exp Cell Res. 1999. 248:30–43.
20. LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998. 17:3247–3259.
21. Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997. 388:300–304.
22. Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998. 58:5315–5320.
23. Hague A, Moorghen M, Hicks D, Chapman M, Paraskeva C. BCL-2 expression in human colorectal adenomas and carcinomas. Oncogene. 1994. 9:3367–3370.
24. Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999. 59:6097–6102.
25. Zhang T, Otevrel T, Gao Z, Gao Z, Ehrlich SM, Fields JZ, et al. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001. 61:8664–8667.
26. Krieg A, Mahotka C, Krieg T, Grabsch H, Müller W, Takeno S, et al. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002. 86:737–743.




 ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download