Abstract
Inherited hemolytic anemias (IHAs) are genetic diseases that present with anemia due to the increased destruction of circulating abnormal RBCs. The RBC abnormalities are classified into the three major disorders of membranopathies, hemoglobinopathies, and enzymopathies. Traditional diagnosis of IHA has been performed via a step-wise process combining clinical and laboratory findings. Nowadays, the etiology of IHA accounts for germline mutations of the responsible genes coding for the structural components of RBCs. Recent advances in molecular technologies, including next-generation sequencing, inspire us to apply these technologies as a first-line approach for the identification of potential mutations and to determine the novel causative genes in patients with IHAs. We herein review the concept and strategy for the genetic diagnosis of IHAs and provide an overview of the preparations for clinical applications of the new molecular technologies.
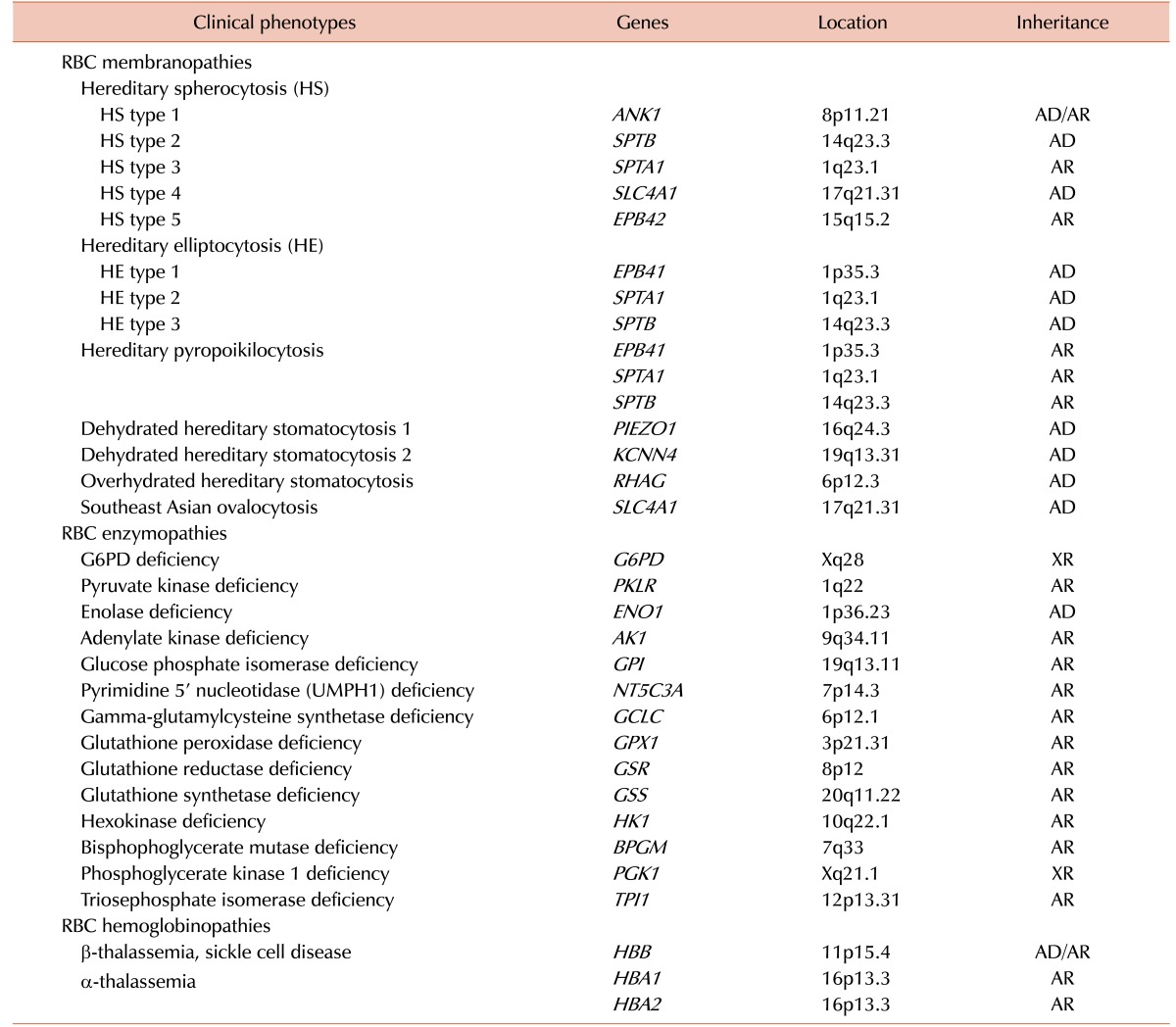
The term inherited hemolytic anemia (IHA) encompasses a diverse group of genetically and phenotypically heterogeneous disorders that result from an increase in the rate of RBC destruction [1]. The severity of the anemia or the course of the onset of hemolysis depends on the extent of this destruction. Mild hemolysis can be asymptomatic, while the anemia in severe hemolysis can be life threatening and cause angina and cardiopulmonary decompensation [23]. IHA develops when the RBCs themselves are defective (intrinsic HA). Defects in hemoglobin (Hb), the RBC membrane, and RBC enzymes are the major causes of intrinsic HA, which are commonly referred to as hemoglobinopathy, membranopathy, and enzymopathy, respectively [456]. Most intrinsic HAs are inherited disorders, but paroxysmal nocturnal hemoglobinuria is exceptional [7]. The most common of these disorders are α- and β-hemoglobinopathies, glucose-6-phosphate dehydrogenase (G6PD) deficiency, and hereditary spherocytosis (HS), which affect millions of people worldwide [8]. A Korean IHA survey-based study reported RBC membranopathies in 64.0%, hemoglobinopathies in 19.9%, and RBC enzymopathies in 13.3% of cases [9]. Traditional diagnosis of IHA has been done via a step-wise process including RBC morphology, membrane protein analysis, Hb electrophoresis, and measurement of RBC enzyme levels [10]. Occasionally, accurate diagnosis of IHA is a challenge because the clinical features may overlap in cases with different etiologies and it is not possible to distinguish between them using conventional diagnostic techniques [1112]. Therefore, there is still a need for more sensitive and reliable diagnostic tests to improve the accuracy of IHA diagnosis. A genome-wide association study of Hb concentration and related parameters (Hb, mean cell hemoglobin [MCH], mean cell hemoglobin concentration [MCHC], mean cell volume [MCV], packed cell volume [PCV], and RBC) has revealed the possible effects of 75 independent genetic loci on the genetic mechanisms and biological pathways that control RBC formation and function [13]. The range of genetic abnormalities has been extensively characterized [14] and genetic testing is available to identify the specific mutation(s) of IHAs (Table 1). Sanger sequencing is widely applied to identify disease-causing mutations in cases where traditional testing has failed or when a patient has been extensively transfused, leading to confounding biochemical and other testing findings due to mixed RBC populations. Recent technological advances including next-generation sequencing (NGS) provide a cost-effective and rapid approach to molecular diagnosis of IHA through extensive and simultaneous evaluation of a group of disease-causing genes [15].
This review describes effective diagnosis of IHA in the current genetic era and introduces recent insights into advanced genotype-phenotype correlation on the basis of genetics; we also discuss the possible implications of NGS in clinical practice for IHA patients.
Knowledge of RBC membrane structure is important because defects in its structure underlie multiple IHAs [16]. The human RBC membrane consists of three basic components: a lipid bilayer, transmembrane linker proteins, and a two-dimensional spectrin-based cytoskeleton network [171819]. Connections of the two layers depend on different linker proteins with binding sites, respectively, for the cytoplasmic domains of the integral membrane proteins (band 3 and glycophorin C) embedded in the lipid bilayer and specific regions of spectrin proteins in the cytoskeleton (Fig. 1). RBC membranopathies are the result of qualitative abnormalities or quantitative deficiencies of the RBC cytoskeletal proteins and can be divided into those resulting from structural protein loss including HS, hereditary elliptocytosis (HE) and hereditary ovalocytosis and membrane transport dysfunction (hereditary stomatocytosis) (Fig. 2) [1620]. Defects that interrupt the vertical structure (spectrin-actin interaction) underlie the biochemical and molecular basis of HS, whereas defects in horizontal interactions (skeletal attachment to membrane proteins) cause HE. All RBC membranopathies share common features, including the loss of surface area, change in morphology, and the resultant tendency for splenic sequestration and extravascular hemolysis resulting in chronic anemia of variable severity [2122]. RBC membranopathies are typically diagnosed by peripheral blood morphology and headed toward confirmation by genetic testing.
HS is an inherited disorder characterized by the presence of spherical-shaped RBCs on peripheral blood smears and is most commonly associated with autosomal dominant inheritance [23]. HS RBC membranes in HS patients show qualitative and/or quantitative abnormalities of proteins including isolated ankyrin or combined ankyrin and spectrin, isolated spectrin, band 3, and protein 4.2. The defects in the membrane components increase membrane fragility and induce vesiculation with or without the band 3 protein. HS is diagnosed through laboratory tests including RBC morphology and osmotic fragility test as well as family history (Fig. 3). The osmotic fragility test has been considered the gold standard screening test for HS but provides false-negative findings in about 25% of patients [24]. Eosin-5'-maleimide measurement and SDS-polyacrylamide gel electrophoresis of erythrocyte membrane proteins are also useful for screening HS, but standardization of these methods is currently lacking [2526]. None of the HS screening tests can detect all patients because the clinical phenotypes are widely variable, ranging from asymptomatic to severely affected [27]. A few patients with mild hemolysis may develop marked anemia if their bone marrow erythrocyte production is transiently halted by viral (parvovirus B19) or other infections [28]. This scenario would be an aplastic crisis since the bone marrow can no longer compensate for ongoing hemolysis. In neonates or transfused individuals, diagnosis can be difficult due to unclear morphological features and screening tests may be unreliable [23]. Molecular testing is useful for primary differential diagnosis or confirming HS and defining the genotype-phenotype correlation [242930]. The defects in RBC membrane components in HS are typically caused by their corresponding gene mutations. Mutations of the ANK1 (ankyrin 1), SPTB (spectrin, beta, erythrocytic), SPTA1 (spectrin, alpha, erythrocytic 1), SLC4A1 (band 3; solute carrier family 4, member 1), and EPB42 (erythrocyte membrane protein band 4.2) genes have been detected in HS patients [313233]. ANK1 mutations (approximately 50%) are the most common cause of HS, followed by mutations in spectrin genes (SPTB: approximately 20% and SPTA1: approximately 5%), SLC4A1 (approximately 15%), and EPB42 (approximately 10%) [21]. In autosomal dominant HS (75%), nonsense and frameshift mutations of ANK1, SLC4A1, and SPTB predominate. Recessive HS is most often due to compound heterozygosity of defects in ANK1, SPTA1, or EPB42 [34]. Most identified mutations are private, which means no frequent defect is found, and nearly every family has a unique mutation. Genetic-based diagnosis of HS in 25 Korean HS patients revealed the ANK1 gene mutation to be the most common, followed by heterozygous SPTB genes. The genotype-phenotype correlations were clarified after a combined analysis of their cases and a literature review. Anemia was most severe in patients with mutations in the spectrin-binding domain of the ANK1 gene. Splenectomy was more frequently performed in patients with ANK1 mutations (32%) than in those with SPTB mutations [35]. As discussed later in this review, the current availability of advanced genomic surveys, such as NGS, allows one to overcome the limitations of the current diagnostic methods and provide additional information to assess the pathogenicity of identified genetic variants by comprehensive genotype-phenotype analyses.
HE is a group of disorders characterized by the presence of elliptical-shaped RBCs [36]. HE has a worldwide distribution but is more common in individuals of African and Mediterranean ancestry [3738]. Most patients are asymptomatic; however, a few neonatal presentations can be dramatic, with jaundice, hemolysis, and hydrops fetalis [39]. In Korea, HE is the cause for 1.4% (6/431) of IHA cases [40] and 15 cases have been reported including a genetically identified family with an SPTA1 mutation [41]. HE is inherited in an autosomal dominant fashion and the majority of HE-associated defects occur due to qualitative and quantitative defects in the RBC membrane skeleton proteins, α-spectrin, β-spectrin, or protein 4.1R. Mutations in SPTA1 are the most common, occurring in 65% of HE cases, followed by mutations in SPTB (30%) and EPB41 (5%) [21]. Interestingly, SPTB mutations identified in HE are located on the tetramerization domain but are distributed between the actin-binding domain and spectrin repeats in HS [35]. Hereditary pyropoikilocytosis (HPP) represents a subtype of common HE as evidenced by the coexistence of both HE and HPP in the same family [42]. Patients with severe HE should be considered for splenectomy; however, some degree of hemolysis persists in post-splenectomy HE patients that indicates an incomplete response to splenectomy [8]. Targeted sequencing by NGS is an efficient approach to identify or confirm the diagnosis of HE and HPP, especially in severe, transfusion-dependent cases where the RBC phenotype cannot be evaluated. In addition, causative molecular diagnosis allows identification of genotype-phenotype correlations in theses heterogeneous disorders and may assist in prognosis determination [43].
The energy for RBCs is dependent upon the production of adenosine triphosphate (ATP) through glycolysis, and ATP is the only source of energy for the RBCs (Fig. 4). Defects in the glycolytic pathway enzymes have been described in metabolic pathways and almost all are associated with chronic HA [44]. Enzymopathies of the pentose phosphate pathway and glutathione metabolism are associated with acute hemolytic crises after exposure to oxidant substances. Deficiencies or malfunctions of these enzymes generally impair cellular energy balance and/or increase the levels of oxidative stress [45]. Since the discovery of G6PD deficiency in 1956 followed by pyruvate kinase (PK) deficiency in 1961, RBC enzymopathies associated with IHA have been extensively investigated [464748]. The mode of inheritance is autosomal recessive for almost all erythroenzymopathies, except for adenosine deaminase overproduction, which is autosomal dominant and G6PD and phosphoglycerate kinase deficiencies, which are X-linked. In contrast to other IHAs, the morphology of the RBCs shows no specific abnormalities. Diagnosis is based on the detection of reduced specific enzyme activity and molecular characterization of the defect at the DNA level [4950].
G6PD deficiency is the most common RBC enzymopathy, affecting 400 million people worldwide [5152] and is especially prevalent in areas of high malaria infection [53]. G6PD deficiency has an X-linked recessive mode of inheritance and most known mutations in the G6PD gene decrease the enzyme stability. Since these cells do not have the ability to efficiently synthesize glutathione and replenish their enzyme levels, the sulfhydryl groups of hemoglobin are oxidized and damage the RBC membrane as cells age during their 120-day lifespan in circulation. The other mechanism is the decreased enzyme activity, in which the diminished ability of RBCs to withstand stress increases the risk of destruction by hemolysis. Biochemical analysis of G6PD enzyme activity levels is commonly used to screen children with unexplained persistent jaundice [5455]. The analysis may also be used to help establish a diagnosis for people with unexplained episodes of HA, jaundice, or dark urine [56]. The World Health Organization groups G6PD deficiency into five categories, in which G6PD enzyme activity of less than 10% is considered a severe deficiency [57]. Sometimes, patients showing nearly normal G6PD activity can be overlooked because molecular analyses of G6PD are only considered for episodes of hemolytic crisis [58]. Various exogenous stressors such as a medication, fava beans, or infection may trigger hemolysis [59]. At least 186 G6PD mutations have been documented, most of which (85%, 159/186) are single nucleotide substitutions leading to missense variants [60]. In Korea, seven genetically identified G6PD deficiencies have been reported. The mutations were not derived from shared ancestor but have arisen by independent mutational events [61]. With respect to genotype-phenotype associations, the frequencies of Class I mutations found in exon 10 are significantly higher than those in other exons in G6PD deficiency [60]. Mutations in this region, which encodes the binding interface between the subunits, have a highly deleterious effect on enzyme activity by disrupting the quaternary structure and stability of the protein and are the most easily identifiable in the general population [6263]. The application of NGS is useful to diagnose the common, rare and novel variants in G6PD deficiency [64].
PK deficiency is a common enzymatic defect of RBCs. Its clinical features are highly variable, ranging from very mild or fully compensated forms to life-threatening neonatal anemia and jaundice necessitating exchange transfusions. PK deficiencies are caused by PKLR mutations and are the most common cause of congenital non-spherocytic HA. Over 200 PKLR mutations have been described [6566]. Up to 70% of PK-deficient alleles carry a missense mutation followed by splicing and stop codon mutations (13% and 5%, respectively; www.lovd.nl/pklr) [50]. Although a genotype-phenotype relationship has not yet been unveiled [67], patients with homozygous null mutations display severe phenotypes including intrauterine growth retardation, severe anemia at birth, and blood transfusion dependence [68]. Splenectomy can reduce the need for transfusion in transfusion-dependent patients, although it does not eliminate hemolysis [44]. Recently, Korean siblings with compound heterozygous null mutations in the PKLR gene have been reported. They initially exhibited congenital dyserythropoietic anemia (CDA)-associated features and Sanger sequencing of the CDA-causing genes was negative. NGS was applied to efficiently identify PKLR mutations as the causal gene in these patients. Although they had a severe phenotype, the patients were eventually cured by hematopoietic stem cell transplantation combined with splenectomy [69].
The hemoglobinopathies are a group of disorders caused by genetic defects that result in the abnormal structure of one of the globin chains of the Hb molecule. With approximately 7% of the worldwide population being carriers, hemoglobinopathies are the most common monogenic diseases and one of the world's major health problems [70]. They fall into two main groups: thalassemia syndromes and structural Hb variants (abnormal hemoglobins). α- and β-thalassemia are the main types of thalassemia; the main structural Hb variants are Hb S, E, and C [71]. α-thalassemia occurs when a gene(s) related to the α-globin protein are deleted or mutated, which occurs most often in persons from Southeast Asia, the Middle East, China, and in those of African descent [72]. β-thalassemia occurs most often in persons of Mediterranean origin and is caused by β-globin gene mutations. To a lesser extent, Chinese, other Asians, and African Americans can also be affected [73]. Some hemoglobinopathies show abnormal RBC morphologies such as target and sickle cells (Fig. 2D, E). Laboratory tests to diagnose hemoglobinopathies are based on the detection of abnormal Hb and include electrophoresis, isoelectric focusing, and high-performance liquid chromatography [7475]. Molecular diagnosis can be made by a variety of techniques, most commonly by Sanger sequencing of the genes coding α- and β-globin and restriction analysis when possible. Over 1,200 genetic alterations that affect the DNA sequence of the human α-like (HBZ, HBA2, HBA1, and HBQ1) and β-like (HBE1, HBG2, HBG1, HBD, and HBB) globin genes are mainly responsible for the observed clinical heterogeneity [76]. Multiplex ligation-dependent probe amplification has become a favored technique to assess for deletions or duplications and can effectively identify different and unknown types of α-globin gene rearrangements to allow the characterization of previously unsolved α-thalassemia genotypes within the α-globin gene region [7778].
Some patients with Hb variants do not show clinical symptoms of HA. They may be detected incidentally by abnormal laboratory findings including falsely low oxygen saturation and glycated hemoglobin (HbA1c)/glucose results [7980]. More than 600 Hb variants have been reported and database records (HbVar, http://globin.bx.psu.edu/hbvar) provide extensive phenotypic descriptions, biochemical and hematological effects, associated pathology, and ethnic occurrence accompanied by mutation frequencies and references [81]. In Korea, 11 Hb variants have been reported [82]. Some have been detected coincidentally while measuring HbA1 levels, including Hb G Coushatta, Hb Queens, Hb Hoshida, and Hb Yamagata [8384]. Genetic analysis can be a more practical application to confirm Hb variants, including novel variants, especially in patients without typical findings. Recent studies demonstrated that NGS could provide a comprehensive assessment of thalassemia screening strategies with its superiority in for both sensitivity and specificity, indicating that NGS is a competitive screening method, especially among populations with a high prevalence of disease [85]. Along with genetic improvement, several gene therapies for the treatment of hemoglobinopathies are currently in clinical trials or under development, including therapies utilizing gene replacement therapy using lentiviruses and the latest gene editing techniques [868788].
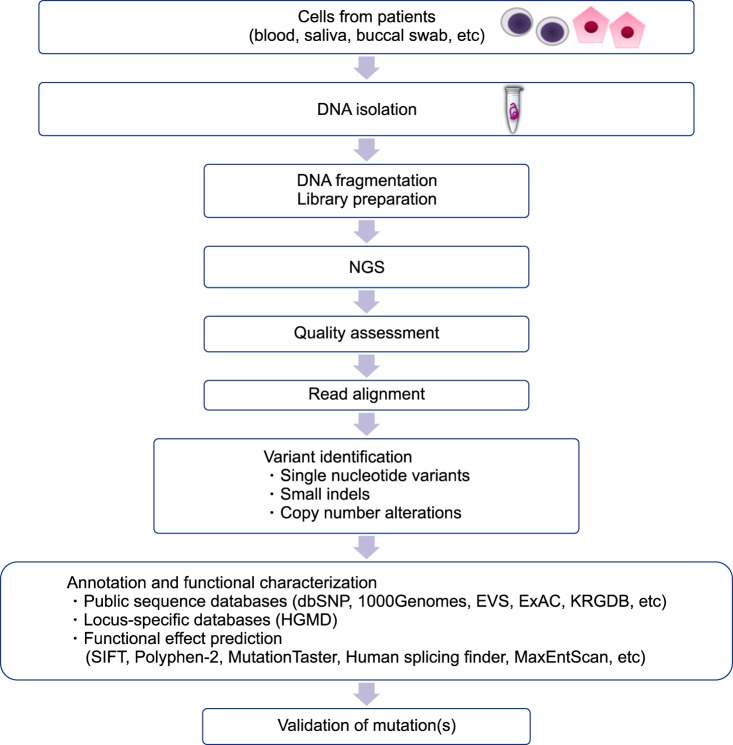
Genetic testing has been used for the confirmatory diagnosis of IHA. Sanger sequencing is primarily performed in order to identify the causative mutations in single gene disorders. It is very lucky to identify mutation(s) in the disease-associated gene in the initial trial. If not, a gene-by-gene approach is required [69]. In these cases, patients may undergo multiple rounds of testing for different genes, a pathway to diagnosis, which can be costly and time-consuming. Additionally, the usefulness of Sanger sequencing is limited for the diagnoses of complex, multi-gene disorders or those with locus heterogeneity. Recent advances in molecular technologies have helped to identify unexpected candidate genes in numerous inherited disorders including IHA [438990]. Various NGS-based methods have been developed, including whole genome sequencing, exome sequencing, and gene panels [91]. Although many recent, successful applications of whole genome sequencing have been reported in establishing the etiology of complex diseases and guiding therapeutic decision-making in neoplastic and nonneoplastic diseases, its use remains challenging and must be carefully evaluated before its clinical implementation as a diagnostic test [92]. Whole-exome sequencing has unveiled numerous causal mutations and genetic modifiers of disease severity in various disorders including IHA [69939495]. Gene panels are currently an attractive option in clinical laboratories because of their reasonable costs, relatively acceptable turn-around time, decreased complexity of data analysis, better coverage over the regions of interest, and reduced incidental findings [969798]. NGS panels consisting of common disease-causing genes have been developed and applied to routine molecular diagnosis for undiagnosed IHA patients and their families [1599]. In particular, patients with the co-presence of membranopathy, enzymopathy, and/or hemoglobinopathy [25100] can be effectively diagnosed using this new technology. Causal gene identification can be deduced through an efficient and reliable strategy to impute and analyze NGS data [101] (Fig. 5). The expanded implementation of the new technology will increase our knowledge of the genetic and genomic differences among individuals, gradually leading to a shift in the clinical management and the therapeutic plan from a population-based approach to a personalized therapy for individual patients [102]. However, it cannot be overemphasized that each laboratory should develop a clinical-grade NGS panel and validate its performance, including analysis and interpretation, before clinical application. A validation strategy is needed to fulfill the requirements set out by the global and national standardized guidelines for NGS panels [103].
Over the years, IHAs caused by RBC membranopathy, RBC enzymopathy, and RBC hemoglobinopathy were screened and diagnosed by using conventional methods including RBC morphology, membrane protein analysis, Hb electrophoresis, and measurement of RBC enzyme levels. In the genetic era, Sanger sequencing became useful for detecting genetic mutations that cause IHAs. Recent advances in genetic technology utilizing NGS enabled us to better identify various genetic mutations that can cause IHAs. The accurate diagnosis of IHA will be feasible by the use of NGS and associated genetic analyses in the clinical laboratory that could overcome the inconclusive and less accurate morphologic and biochemical analysis. We believe that understanding IHA on genetic basis and applying genetic technologies for routine clinical laboratory testing will improve the accuracy and efficiency of IHA diagnosis and give us insights for precision medicine of each affected individual.
ACKNOWLEDGMENTS
We would like to thank our colleague, Jaewoong Lee for providing illustration supports in this study.
References
2. Ucar K. Clinical presentation and management of hemolytic anemias. Oncology (Williston Park). 2002; 16(9 Suppl 10):163–170. PMID: 12380967.
3. Lode HN, Krings G, Schulze-Neick I, et al. Pulmonary hypertension in a case of Hb-Mainz hemolytic anemia. J Pediatr Hematol Oncol. 2007; 29:173–177. PMID: 17356397.

4. Chiu D, Lubin B. Oxidative hemoglobin denaturation and RBC destruction: the effect of heme on red cell membranes. Semin Hematol. 1989; 26:128–135. PMID: 2658088.
5. Jacobasch G, Rapoport SM. Hemolytic anemias due to erythrocyte enzyme deficiencies. Mol Aspects Med. 1996; 17:143–170. PMID: 8813716.
6. Zanella A, Fermo E, Bianchi P, Valentini G. Red cell pyruvate kinase deficiency: molecular and clinical aspects. Br J Haematol. 2005; 130:11–25. PMID: 15982340.

7. Parker CJ. Paroxysmal nocturnal hemoglobinuria. Curr Opin Hematol. 2012; 19:141–148. PMID: 22395662.

8. Gallagher PG. Diagnosis and management of rare congenital nonimmune hemolytic disease. Hematology Am Soc Hematol Educ Program. 2015; 2015:392–399. PMID: 26637748.

9. Park ES, Jung HL, Kim HJ, et al. Hereditary hemolytic anemia in Korea from 2007 to 2011: A study by the Korean Hereditary Hemolytic Anemia Working Party of the Korean Society of Hematology. Blood Res. 2013; 48:211–216. PMID: 24086942.

10. Barcellini W, Fattizzo B. Clinical applications of hemolytic markers in the differential diagnosis and management of hemolytic anemia. Dis Markers. 2015; 2015:635670. PMID: 26819490.

11. King MJ, Zanella A. Hereditary red cell membrane disorders and laboratory diagnostic testing. Int J Lab Hematol. 2013; 35:237–243. PMID: 23480868.

12. Christensen RD, Nussenzveig RH, Yaish HM, Henry E, Eggert LD, Agarwal AM. Causes of hemolysis in neonates with extreme hyperbilirubinemia. J Perinatol. 2014; 34:616–619. PMID: 24762414.

13. van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012; 492:369–375. PMID: 23222517.
14. Sankaran VG, Gallagher PG. Applications of high-throughput DNA sequencing to benign hematology. Blood. 2013; 122:3575–3582. PMID: 24021670.

15. Agarwal AM, Nussenzveig RH, Reading NS, et al. Clinical utility of next-generation sequencing in the diagnosis of hereditary haemolytic anaemias. Br J Haematol. 2016; 174:806–814. PMID: 27292444.

16. Andolfo I, Russo R, Gambale A, Iolascon A. New insights on hereditary erythrocyte membrane defects. Haematologica. 2016; 101:1284–1294. PMID: 27756835.

17. Shohet SB, Bicknese SE. Defining the architecture of the red blood cell membrane: newer biophysical approaches. Am J Hematol. 1993; 42:19–24. PMID: 8416291.

18. Kusumi A, Sako Y. Cell surface organization by the membrane skeleton. Curr Opin Cell Biol. 1996; 8:566–574. PMID: 8791449.

19. Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood. 2008; 112:3939–3948. PMID: 18988878.

20. Palek J. Introduction: red blood cell membrane proteins, their genes and mutations. Semin Hematol. 1993; 30:1–3.
21. An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008; 141:367–375. PMID: 18341630.

22. Gallagher PG. Update on the clinical spectrum and genetics of red blood cell membrane disorders. Curr Hematol Rep. 2004; 3:85–91. PMID: 14965483.
23. Bolton-Maggs PH, Langer JC, Iolascon A, Tittensor P, King MJ. General Haematology Task Force of the British Committee for Standards in Haematology. Guidelines for the diagnosis and management of hereditary spherocytosis-2011 update. Br J Haematol. 2012; 156:37–49. PMID: 22055020.
24. Bianchi P, Fermo E, Vercellati C, et al. Diagnostic power of laboratory tests for hereditary spherocytosis: a comparison study in 150 patients grouped according to molecular and clinical characteristics. Haematologica. 2012; 97:516–523. PMID: 22058213.

25. King MJ, Garçon L, Hoyer JD, et al. ICSH guidelines for the laboratory diagnosis of nonimmune hereditary red cell membrane disorders. Int J Lab Hematol. 2015; 37:304–325. PMID: 25790109.

26. King MJ, Behrens J, Rogers C, Flynn C, Greenwood D, Chambers K. Rapid flow cytometric test for the diagnosis of membrane cytoskeleton-associated haemolytic anaemia. Br J Haematol. 2000; 111:924–933. PMID: 11122157.

27. Farias MG. Advances in laboratory diagnosis of hereditary spherocytosis. Clin Chem Lab Med. 2017; 55:944–948. PMID: 27837594.

28. Frickhofen N, Chen ZJ, Young NS, Cohen BJ, Heimpel H, Abkowitz JL. Parvovirus B19 as a cause of acquired chronic pure red cell aplasia. Br J Haematol. 1994; 87:818–824. PMID: 7986722.

29. Hassoun H, Vassiliadis JN, Murray J, et al. Characterization of the underlying molecular defect in hereditary spherocytosis associated with spectrin deficiency. Blood. 1997; 90:398–406. PMID: 9207476.
30. Mariani M, Barcellini W, Vercellati C, et al. Clinical and hematologic features of 300 patients affected by hereditary spherocytosis grouped according to the type of the membrane protein defect. Haematologica. 2008; 93:1310–1317. PMID: 18641031.

31. Delaunay J. The molecular basis of hereditary red cell membrane disorders. Blood Rev. 2007; 21:1–20. PMID: 16730867.

32. Iolascon A, Miraglia del Giudice E, Perrotta S, Alloisio N, Morlé L, Delaunay J. Hereditary spherocytosis: from clinical to molecular defects. Haematologica. 1998; 83:240–257. PMID: 9573679.
33. Eber SW, Gonzalez JM, Lux ML, et al. Ankyrin-1 mutations are a major cause of dominant and recessive hereditary spherocytosis. Nat Genet. 1996; 13:214–218. PMID: 8640229.

34. Eber S, Lux SE. Hereditary spherocytosis--defects in proteins that connect the membrane skeleton to the lipid bilayer. Semin Hematol. 2004; 41:118–141. PMID: 15071790.

35. Park J, Jeong DC, Yoo J, et al. Mutational characteristics of ANK1 and SPTB genes in hereditary spherocytosis. Clin Genet. 2016; 90:69–78. PMID: 26830532.
36. Da Costa L, Galimand J, Fenneteau O, Mohandas N. Hereditary spherocytosis, elliptocytosis, and other red cell membrane disorders. Blood Rev. 2013; 27:167–178. PMID: 23664421.

37. Glele-Kakai C, Garbarz M, Lecomte MC, et al. Epidemiological studies of spectrin mutations related to hereditary elliptocytosis and spectrin polymorphisms in Benin. Br J Haematol. 1996; 95:57–66. PMID: 8857939.
38. Dhermy D, Schrével J, Lecomte MC. Spectrin-based skeleton in red blood cells and malaria. Curr Opin Hematol. 2007; 14:198–202. PMID: 17414207.

39. Gallagher PG, Weed SA, Tse WT, et al. Recurrent fatal hydrops fetalis associated with a nucleotide substitution in the erythrocyte beta-spectrin gene. J Clin Invest. 1995; 95:1174–1182. PMID: 7883966.

40. Cho HS, Hah JO, Kang IJ, et al. Hereditary hemolytic anemia in Korea: a retrospective study from 1997 to 2006. Korean J Hematol. 2007; 42:197–205.

41. Han E, Kim A, Park J, et al. Spectrin Tunis (Sp alpha (I/78)) in a Korean family with hereditary elliptocytosis. Ann Lab Med. 2013; 33:386–389. PMID: 24003435.

42. Hoffman R, Benz EJ Jr, Silberstein LE, Heslop H, Weitz J, Anastasi J. Hematology: basic principles and practice. 6th ed. PA: Elsevier Saunders;2013. p. 418–426.
43. Niss O, Chonat S, Dagaonkar N, et al. Genotype-phenotype correlations in hereditary elliptocytosis and hereditary pyropoikilocytosis. Blood Cells Mol Dis. 2016; 61:4–9. PMID: 27667160.

44. Prchal JT, Gregg XT. Red cell enzymes. Hematology Am Soc Hematol Educ Program. 2005; 19–23. PMID: 16304354.

45. Koralkova P, van Solinge WW, van Wijk R. Rare hereditary red blood cell enzymopathies associated with hemolytic anemia - pathophysiology, clinical aspects, and laboratory diagnosis. Int J Lab Hematol. 2014; 36:388–397. PMID: 24750686.

46. Vives i Corrons JL. Chronic non-spherocytic haemolytic anaemia due to congenital pyrimidine 5' nucleotidase deficiency: 25 years later. Baillieres Best Pract Res Clin Haematol. 2000; 13:103–118. PMID: 10916681.
47. Kahn A, Kaplan JC, Dreyfus JC. Advances in hereditary red cell enzyme anomalies. Hum Genet. 1979; 50:1–27. PMID: 157322.

48. Fiorelli G, Martinez di Montemuros F, Cappellini MD. Chronic non-spherocytic haemolytic disorders associated with glucose-6-phosphate dehydrogenase variants. Baillieres Best Pract Res Clin Haematol. 2000; 13:39–55. PMID: 10916677.

49. Zanella A, Fermo E, Bianchi P, Chiarelli LR, Valentini G. Pyruvate kinase deficiency: the genotype-phenotype association. Blood Rev. 2007; 21:217–231. PMID: 17360088.

50. Canu G, De Bonis M, Minucci A, Capoluongo E. Red blood cell PK deficiency: An update of PK-LR gene mutation database. Blood Cells Mol Dis. 2016; 57:100–109. PMID: 26832193.

51. Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008; 371:64–74. PMID: 18177777.

52. Howes RE, Battle KE, Satyagraha AW, Baird JK, Hay SI. G6PD deficiency: global distribution, genetic variants and primaquine therapy. Adv Parasitol. 2013; 81:133–201. PMID: 23384623.
53. von Seidlein L, Auburn S, Espino F, et al. Review of key knowledge gaps in glucose-6-phosphate dehydrogenase deficiency detection with regard to the safe clinical deployment of 8-aminoquinoline treatment regimens: a workshop report. Malar J. 2013; 12:112. PMID: 23537118.

54. Keihanian F, Basirjafari S, Darbandi B, et al. Comparison of quantitative and qualitative tests for glucose-6-phosphate dehydrogenase deficiency in the neonatal period. Int J Lab Hematol. 2017; 39:251–260. PMID: 28258653.

55. Nadarajan V, Shanmugam H, Sthaneshwar P, et al. Modification to reporting of qualitative fluorescent spot test results improves detection of glucose-6-phosphate dehydrogenase (G6PD)-deficient heterozygote female newborns. Int J Lab Hematol. 2011; 33:463–470. PMID: 21501392.

56. Minucci A, Giardina B, Zuppi C, et al. Glucose-6-phosphate dehydrogenase laboratory assay: How, when, and why? IUBMB Life. 2009; 61:27–34. PMID: 18942156.

57. Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ. 1989; 67:601–611. PMID: 2633878.
58. Vaca G, Arámbula E, Monsalvo A, et al. Glucose-6-phosphate dehydrogenase (G-6-PD) mutations in Mexico: four new G-6-PD variants. Blood Cells Mol Dis. 2003; 31:112–120. PMID: 12850494.

59. Domingo GJ, Satyagraha AW, Anvikar A, et al. G6PD testing in support of treatment and elimination of malaria: recommendations for evaluation of G6PD tests. Malar J. 2013; 12:391. PMID: 24188096.

60. Minucci A, Moradkhani K, Hwang MJ, Zuppi C, Giardina B, Capoluongo E. Glucose-6-phosphate dehydrogenase (G6PD) mutations database: review of the "old" and update of the new mutations. Blood Cells Mol Dis. 2012; 48:154–165. PMID: 22293322.

61. Lee J, Park J, Choi H, et al. Genetic profiles of Korean patients with glucose-6-phosphate dehydrogenase deficiency. Ann Lab Med. 2017; 37:108–116. PMID: 28028996.

62. Gómez-Manzo S, Marcial-Quino J, Vanoye-Carlo A, et al. Glucose-6-phosphate dehydrogenase: update and analysis of new mutations around the world. Int J Mol Sci. 2016; 17:E2069. PMID: 27941691.

63. Luzzatto L, Nannelli C, Notaro R. Glucose-6-phosphate dehydrogenase deficiency. Hematol Oncol Clin North Am. 2016; 30:373–393. PMID: 27040960.

64. Bogari NM. Next generation sequencing (NGS) in glucose-6-phosphate dehydrogenase (G6PD) deficiency studies. Bioinformation. 2016; 12:41–43. PMID: 28104958.

66. Climent F, Roset F, Repiso A, Pérez de la Ossa P. Red cell glycolytic enzyme disorders caused by mutations: an update. Cardiovasc Hematol Disord Drug Targets. 2009; 9:95–106. PMID: 19519368.

67. Grace RF, Zanella A, Neufeld EJ, et al. Erythrocyte pyruvate kinase deficiency: 2015 status report. Am J Hematol. 2015; 90:825–830. PMID: 26087744.

68. Rider NL, Strauss KA, Brown K, et al. Erythrocyte pyruvate kinase deficiency in an old-order Amish cohort: longitudinal risk and disease management. Am J Hematol. 2011; 86:827–834. PMID: 21815188.

69. Kim M, Park J, Lee J, et al. Hemolytic anemia with null PKLR mutations identified using whole exome sequencing and cured by hematopoietic stem cell transplantation combined with splenectomy. Bone Marrow Transplant. 2016; 51:1605–1608. PMID: 27595284.

70. Weatherall DJ. Hemoglobinopathies worldwide: present and future. Curr Mol Med. 2008; 8:592–599. PMID: 18991645.

71. Wang HC, Hsieh LL, Liu YC, et al. The epidemiologic transition of thalassemia and associated hemoglobinopathies in southern Taiwan. Ann Hematol. 2017; 96:183–188. PMID: 27891555.

72. Higgs DR, Weatherall DJ. The alpha thalassaemias. Cell Mol Life Sci. 2009; 66:1154–1162. PMID: 19020805.

73. Olivieri NF. The beta-thalassemias. N Engl J Med. 1999; 341:99–109. PMID: 10395635.
74. Clarke GM, Higgins TN. Laboratory investigation of hemoglobinopathies and thalassemias: review and update. Clin Chem. 2000; 46:1284–1290. PMID: 10926923.

75. Kutlar F. Diagnostic approach to hemoglobinopathies. Hemoglobin. 2007; 31:243–250. PMID: 17486507.

76. Patrinos GP, Kollia P, Papadakis MN. Molecular diagnosis of inherited disorders: lessons from hemoglobinopathies. Hum Mutat. 2005; 26:399–412. PMID: 16138310.

77. Colosimo A, Gatta V, Guida V, et al. Application of MLPA assay to characterize unsolved a-globin gene rearrangements. Blood Cells Mol Dis. 2011; 46:139–144. PMID: 21190870.
78. Kipp BR, Roellinger SE, Lundquist PA, Highsmith WE, Dawson DB. Development and clinical implementation of a combination deletion PCR and multiplex ligation-dependent probe amplification assay for detecting deletions involving the human a-globin gene cluster. J Mol Diagn. 2011; 13:549–557. PMID: 21708285.
79. Higgins T. Hemoglobinopathies/thalassemias: Why clinical biochemists need to know about them. Clin Biochem. 2017; [Epub ahead of print].

81. Giardine B, Borg J, Viennas E, et al. Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 2014; 42:D1063–D1069. PMID: 24137000.

82. Jo I, Jang W, Chae H, et al. Hemoglobin Kansas: first Korean family and literature review. Ann Lab Med. 2017; 37:352–354. PMID: 28445020.

83. Lee ST, Kim MS, Choi DY, Kim SK, Ki CS. Incidence of variant hemoglobin (Hb) and increased fetal Hb concentrations and their effect on Hb A1c measurement in a Korean population. Clin Chem. 2006; 52:1445–1446. PMID: 16798979.

84. Jung CL, Kwon KJ, Hong KS, et al. Hemoglobin Yamagata: hemoglobin variant detected by HbA1c test. Korean J Lab Med. 2009; 29:536–540. PMID: 20046085.

85. He J, Song W, Yang J, et al. Next-generation sequencing improves thalassemia carrier screening among premarital adults in a high prevalence population: the Dai nationality, China. Genet Med. 2017; [Epub ahead of print].

86. Goodman MA, Malik P. The potential of gene therapy approaches for the treatment of hemoglobinopathies: achievements and challenges. Ther Adv Hematol. 2016; 7:302–315. PMID: 27695619.

87. Traxler EA, Yao Y, Wang YD, et al. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016; 22:987–990. PMID: 27525524.

88. Rai P, Malik P. Gene therapy for hemoglobin disorders - a mini-review. J Rare Dis Res Treat. 2016; 1:25–31. PMID: 27891535.

89. Jamwal M, Aggarwal A, Das A, et al. Next-generation sequencing unravels homozygous mutation in glucose-6-phosphate isomerase, GPIc.1040G>A (p.Arg347His) causing hemolysis in an Indian infant. Clin Chim Acta. 2017; 468:81–84. PMID: 28223188.
90. Del Orbe Barreto R, Arrizabalaga B, De la Hoz AB, et al. Detection of new pathogenic mutations in patients with congenital haemolytic anaemia using next-generation sequencing. Int J Lab Hematol. 2016; 38:629–638. PMID: 27427187.

91. Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015; 17:444–451. PMID: 25232854.

92. Chrystoja CC, Diamandis EP. Whole genome sequencing as a diagnostic test: challenges and opportunities. Clin Chem. 2014; 60:724–733. PMID: 24227285.

93. Han JH, Kim S, Jang H, et al. Identification of a novel p.Q1772X ANK1 mutation in a Korean family with hereditary spherocytosis. PLoS One. 2015; 10:e0131251. PMID: 26107955.

94. Lacy JN, Ulirsch JC, Grace RF, et al. Exome sequencing results in successful diagnosis and treatment of a severe congenital anemia. Cold Spring Harb Mol Case Stud. 2016; 2:a000885. PMID: 27551681.

95. Lettre G. The search for genetic modifiers of disease severity in the β-hemoglobinopathies. Cold Spring Harb Perspect Med. 2012; 2:pii:a015032.

96. Wooderchak-Donahue WL, O'Fallon B, Furtado LV, et al. A direct comparison of next generation sequencing enrichment methods using an aortopathy gene panel- clinical diagnostics perspective. BMC Med Genomics. 2012; 5:50. PMID: 23148498.

97. Dames S, Chou LS, Xiao Y, et al. The development of next-generation sequencing assays for the mitochondrial genome and 108 nuclear genes associated with mitochondrial disorders. J Mol Diagn. 2013; 15:526–534. PMID: 23665194.

98. Sun Y, Ruivenkamp CA, Hoffer MJ, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat. 2015; 36:648–655. PMID: 25772376.

99. Roy NB, Wilson EA, Henderson S, et al. A novel 33-Gene targeted resequencing panel provides accurate, clinical-grade diagnosis and improves patient management for rare inherited anaemias. Br J Haematol. 2016; 175:318–330. PMID: 27432187.

100. Charoenkwan P, Natesirinilkul R, Choeyprasert W, Kulsumritpon N, Sangiamporn O. Coinheritance of hereditary elliptocytosis and deletional hemoglobin H disease. J Pediatr Hematol Oncol. 2017; 39:e69–e70. PMID: 28060122.

101. Hoffmann TJ, Witte JS. Strategies for imputing and analyzing rare variants in association studies. Trends Genet. 2015; 31:556–563. PMID: 26450338.

102. Russo R, Andolfo I, Iolascon A. Next generation research and therapy in red blood cell diseases. Haematologica. 2016; 101:515–517. PMID: 27132276.

103. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–424. PMID: 25741868.

Fig. 1
A schematic representation of red blood cell (RBC) membrane structure with major functional components. The RBC membrane consists of three basic components: a lipid bilayer, transmembrane proteins, and a cytoskeletal network. The major transmembrane proteins are glycoproteins, band 3, and glycophorin. The most abundant protein in the membrane skeleton is spectrin, which is tethered to the phospholipid membrane.
Abbreviations: 4.1, protein band 4.1; 4.2, protein band 4.2; GLUT1, glucose transporter 1; GPA, glycophorin A; GPC, glycophorin C; Rh, rhesus polypeptide; RhAG, Rh-associated glycoprotein.
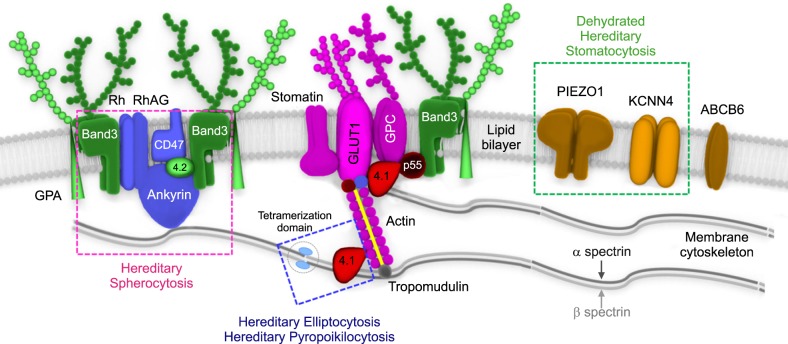
Fig. 2
Peripheral blood smear of inherited hemolytic anemia. (A) Hereditary spherocytosis, (B) hereditary elliptocytosis, (C) hereditary stomatocytosis, (D) β-thalassemia, (E) sickle cell anemia.
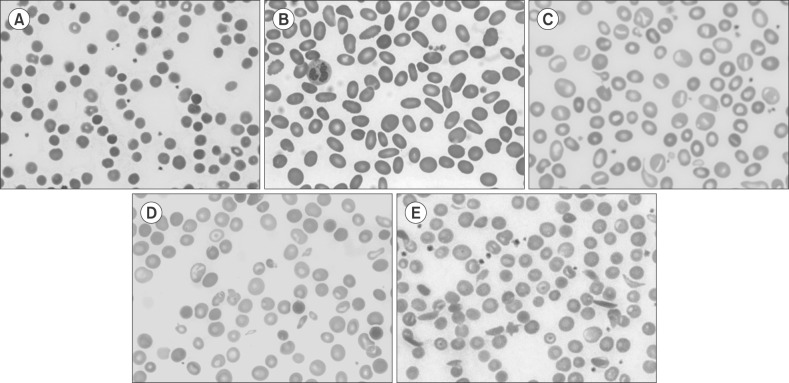
Fig. 3
Stepwise process for genetic-based diagnosis of hereditary spherocytosis.
Abbreviations: CBC, complete blood cell counting; HS, hereditary spherocytosis; LDH, lactate dehydrogenase; NGS, next-generation sequencing; RBC, red blood cell.
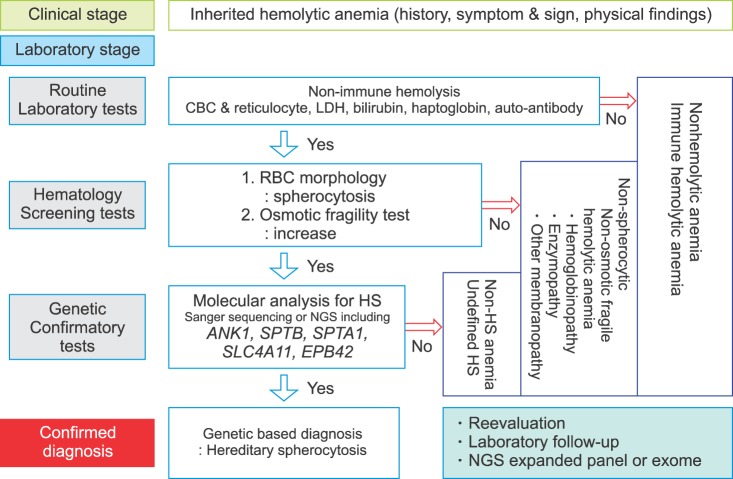
Fig. 4
Anaerobic glycolysis and antioxidant metabolic pathways of red blood cells.
Abbreviations: BPG, bisphosphoglyceric acid; DHAP, dihydroxyacetone phosphate; F6P, fructose 6-phosphate; FDP, fructose-1,6-diphosphate; G3P, glycerol 3-phosphate; G3PD, glyceraldehyde 3-phosphate dehydrogenase; G6P, glucose 6-phosphate; G6PD, glucose-6-phosphate dehydrogenase; GCS, glutamylcysteine synthetase; GPI, glucose-phosphate isomerase; GS, glutathione synthetase; GSH, glutathione; GSSG, glutathione disulfide; HK, hexokinase; LD, lactate dehydrogenase; NADP, nicotinamide adenine dinucleotide phosphate; PEP, phosphoenolpyruvic acid; PFK, phosphofructokinase; PG, phosphoglyceric acid; PGK, phosphoglycerate kinase; PK, pyruvate kinase; Ru5P, ribose-5-phosphate isomerase.
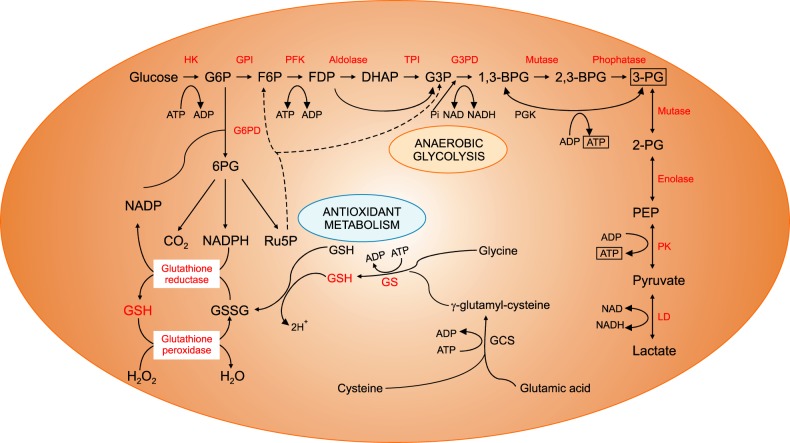
Fig. 5
Overview of steps in the generation of NGS data and analysis.
Abbreviations: NGS, next-generation sequencing; dbSNP, NCBI dbSNP Build 141, http://www.ncbi.nlm.nih.gov/projects/SNP/; 1000Genomes, 1000 Genomes Project, http://www.1000genomes.org/; EVS, Exome Variant Server, http://evs.gs.washington.edu/EVS/; ExAC, Exome Aggregation Consortium database, http://exac.broadinstitute.org/; KRDGB, Korean Reference Genome Database, http://152.99.75.168/KRGDB/menuPages/intro.jsp; SIFT, http://sift.jcvi.org/; PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/; MutationTaster, http://www.mutationtaster.org/; Human splicing findinger, http://www.umd.be/HSF/; MaxEntScan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download