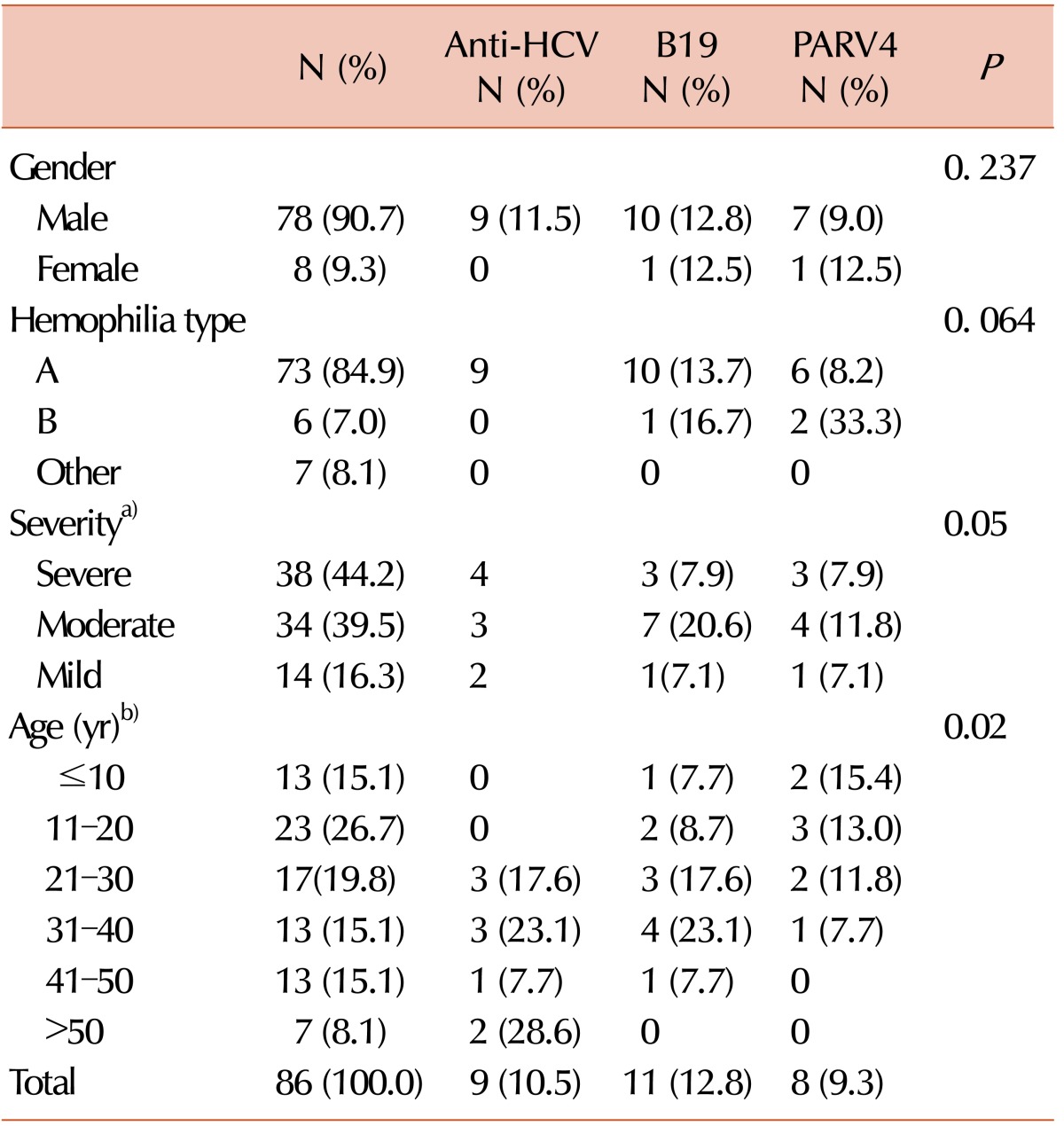
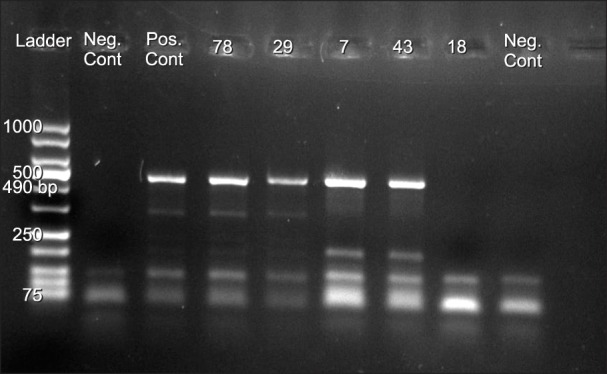
1. Arabzadeh SA, Alizadeh F, Tavakoli A, et al. Human parvovirus B19 in patients with beta thalassemia major from Tehran, Iran. Blood Res. 2017; 52:50–54. PMID:
28401102.

2. Servant A, Laperche S, Lallemand F, et al. Genetic diversity within human erythroviruses: identification of three genotypes. J Virol. 2002; 76:9124–9134. PMID:
12186896.

3. Heegaard ED, Brown KE. Human parvovirus B19. Clin Microbiol Rev. 2002; 15:485–505. PMID:
12097253.

4. Shabani Z, Esghaei M, Keyvani H, et al. Relation between parvovirus B19 infection and fetal mortality and spontaneous abortion. Med J Islam Repub Iran. 2015; 29:197. PMID:
26157715.
5. Ekman A, Hokynar K, Kakkola L, et al. Biological and immunological relations among human parvovirus B19 genotypes 1 to 3. J Virol. 2007; 81:6927–6935. PMID:
17409158.

6. Soucie JM, Siwak EB, Hooper WC, Evatt BL, Hollinger FB. Universal Data Collection Project Working Group. Human parvovirus B19 in young male patients with hemophilia A: associations with treatment product exposure and joint range-of-motion limitation. Transfusion. 2004; 44:1179–1185. PMID:
15265122.

7. Mortimer PP, Luban NL, Kelleher JF, Cohen BJ. Transmission of serum parvovirus-like virus by clotting-factor concentrates. Lancet. 1983; 2:482–484. PMID:
6136645.

8. Rubinstein R, Karabus CD, Smuts H, Kolia F, Van Rensburg EJ. Prevalence of human parvovirus B19 and TT virus in a group of young haemophiliacs in South Africa. Haemophilia. 2000; 6:93–97. PMID:
10781195.

9. Röhrer C, Gärtner B, Sauerbrei A, et al. Seroprevalence of parvovirus B19 in the German population. Epidemiol Infect. 2008; 136:1564–1575. PMID:
18198003.
10. Schmidt I, Blümel J, Seitz H, Willkommen H, Löwer J. Parvovirus B19 DNA in plasma pools and plasma derivatives. Vox Sang. 2001; 81:228–235. PMID:
11903998.

11. Kishore J, Misra R, Gupta D, Ayyagari A. Raised IgM antibodies to parvovirus B19 in juvenile rheumatoid arthritis. Indian J Med Res. 1998; 107:15–18. PMID:
9529776.
12. Soucie JM, De Staercke C, Monahan PE, et al. Evidence for the transmission of parvovirus B19 in patients with bleeding disorders treated with plasma-derived factor concentrates in the era of nucleic acid test screening. Transfusion. 2013; 53:1217–1225. PMID:
22998193.

13. Zakrzewska K, Azzi A, De Biasi E, et al. Persistence of parvovirus B19 DNA in synovium of patients with haemophilic arthritis. J Med Virol. 2001; 65:402–407. PMID:
11536251.

14. Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005; 79:8230–8236. PMID:
15956568.

15. Fryer JF, Kapoor A, Minor PD, Delwart E, Baylis SA. Novel parvovirus and related variant in human plasma. Emerg Infect Dis. 2006; 12:151–154. PMID:
16494735.

16. Simmonds P, Douglas J, Bestetti G, et al. A third genotype of the human parvovirus PARV4 in sub-Saharan Africa. J Gen Virol. 2008; 89:2299–2302. PMID:
18753240.

17. Yu X, Wang J, Zhao B, Ghildyal R. PARV4 co-infection is associated with disease progression in HBV patients in Shanghai. J Med Diagn Methods. 2015; 4:2.
18. Lurcharchaiwong W, Chieochansin T, Payungporn S, Theamboonlers A, Poovorawan Y. Parvovirus 4 (PARV4) in serum of intravenous drug users and blood donors. Infection. 2008; 36:488–491. PMID:
18759058.

19. Benjamin LA, Lewthwaite P, Vasanthapuram R, et al. Human parvovirus 4 as potential cause of encephalitis in children, India. Emerg Infect Dis. 2011; 17:1484–1487. PMID:
21801629.

20. Toppinen M, Norja P, Aaltonen LM, et al. A new quantitative PCR for human parvovirus B19 genotypes. J Virol Methods. 2015; 218:40–45. PMID:
25794796.

21. Tuke PW, Parry RP, Appleton H. Parvovirus PARV4 visualization and detection. J Gen Virol. 2010; 91:541–544. PMID:
19846677.

22. Franchini M, Mannucci PM. Past, present and future of hemophilia: a narrative review. Orphanet J Rare Dis. 2012; 7:24. PMID:
22551339.

23. Marano G, Vaglio S, Pupella S, et al. Human Parvovirus B19 and blood product safety: a tale of twenty years of improvements. Blood Transfus. 2015; 13:184–196. PMID:
25849894.
24. Simmons R, Sharp C, McClure CP, et al. Parvovirus 4 infection and clinical outcome in high-risk populations. J Infect Dis. 2012; 205:1816–1820. PMID:
22492853.

25. Slavov SN, Kashima S, Rocha-Junior MC, et al. Frequent human parvovirus B19 DNA occurrence and high seroprevalence in haemophilic patients from a non-metropolitan blood centre, Brazil. Transfus Med. 2014; 24:130–132. PMID:
24684574.
26. Nishida Y, Arai M, Yamamoto Y, Fukutake K. Serological and virological markers of human parvovirus B19 infection in patients with haemophilia. Haemophilia. 1997; 3:137–142. PMID:
27214724.

27. Shooshtari MM, Foroghi MN, Hamkar R. High prevalence of parvovirus B19 IgG antibody among hemophilia patients in center for special diseases, Shiraz, Iran. Iran J Public Health. 2005; 34:51–54.
28. Sharp CP, Lail A, Donfield S, et al. High frequencies of exposure to the novel human parvovirus PARV4 in hemophiliacs and injection drug users, as detected by a serological assay for PARV4 antibodies. J Infect Dis. 2009; 200:1119–1125. PMID:
19691429.

29. Sharp CP, Lail A, Donfield S, Gomperts ED, Simmonds P. Virologic and clinical features of primary infection with human parvovirus 4 in subjects with hemophilia: frequent transmission by virally inactivated clotting factor concentrates. Transfusion. 2012; 52:1482–1489. PMID:
22043925.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download