Abstract
Background
Mesenchymal stem cells (MSCs) play an important role in hematopoietic stem cell (HSC) maintenance, proliferation, and apoptosis. DNA methyltransferase 1 (DNMT1) is considered an essential factor in the maintenance of HSCs in mammalian cells. Therefore, this study was conducted to evaluate the mRNA expression level of DNMT1 during cord blood (CB)-HSC ex vivo expansion with MSCs.
Methods
Ex vivo cultures of CB-HSCs were performed in three culture conditions for 7 days: cytokines, cytokines with MSCs, and only MSCs. Total and viable cell numbers were counted after 5 and 7 days using trypan blue stain, and the stem cell percentage was then evaluated by flow cytometry. Moreover, in vitro colony-forming unit assay was carried out to detect clonogenic potential of HSCs at days 0 and 7 using MethoCult H4434. Finally, DNMT1 mRNA expression level was evaluated by real-time polymerase chain reaction.
Results
Maximum CB-CD34+ cell expansion was observed on day 7 in all the three cultures. After 7 days, ex vivo expansion of CB-CD34+ cells indicated a significant decrease in DNMT1 expression in the cytokine cultures, whereas in the two co-culture conditions DNMT1 expression was increased. A significant difference between the number of CD34+ and CD34− cells in the cytokine co-culture system was observed.
Ex vivo expansion of cord blood-hematopoietic stem cells (CB-HSCs) is the best way to increase the number of CD34+ cells and improve the kinetics of HSC engraftment [12]. HSC differentiation to precursor cells during ex vivo expansion leads to decreased numbers of primitive HSCs [34]. During normal hematopoiesis, a balance between cell loss (apoptosis and differentiation) and cell gain (proliferation and mitosis) can determine HSC population size [5]. Epigenetic modifications play a critical role in the self-renewal and differentiation of HSCs [67]. DNA methylation of CpG nucleotides is catalyzed by a family of DNA methyltransferase (DNMT) enzymes: DNMT1, DNMT3a, and DNMT3b [89]. DNMT1 is the most abundant DNMT in mammalian cells and is considered to be the key maintenance methyltransferase [10]. DNMT1 regulates distinct patterns of methylation and expression of discrete gene families in long-term HSCs and multipotent and lineage-restricted progenitors [11]. DNMT1 also plays a pivotal role in self-renewal, niche retention, and differentiation of primitive HSCs [12,13]. In our previous study, we demonstrated that mesenchymal stem cells (MSCs) play an important role in hematopoietic regulation by homing and maintaining HSCs in the quiescent phase [14]; however, the precise mechanism by which MSCs regulate HSC differentiation requires further investigation. In this study, DNMT1 mRNA expression levels of cord blood-derived CD34+ cells expanded in co-culture condition with MSCs, in the presence or absence of cytokines, were compared with those cultured with cytokines alone. We also separated the cultured HSCs into CD34+ and CD34− fractions and compared their DNMT1 expression levels with that of freshly isolated HSCs.
Three fresh human umbilical cord blood samples were obtained from healthy full-term pregnant women after obtaining informed consent, according to the Ethical Committee Guidelines of Iranian Blood Transfusion Organization (IBTO). This study was approved by the Medical Ethics Committee of Research Center of IBTO. CD34+ HSCs were isolated using MACS (Miltenyi Biotec, Bergisch Gladbach, Germany). Briefly, the CD34+ cells were labeled with CD34 MicroBeads. The magnetically labeled CD34+ cells were retained within the magnetic column. After removing the column from the magnetic field, the magnetically retained CD34+ cells were eluted as the positively selected cell fraction.
Three heparinized bone marrow samples were obtained from healthy donors according to the Shariati BMT (Bone Marrow Transplantation) Center guidelines. All volunteers were asked to sign an informed consent form prior to bone marrow collection. The bone marrow was diluted using an MSC-specific medium and then isolated by Ficoll-Hypaque density gradient centrifugation. Human bone marrow-derived MSCs were maintained in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% fetal bovine serum. The medium was replaced at least twice a week and non-adherent cells were discarded. MSCs were assayed for CD34, CD45, CD90, CD105, and CD166 expression according to the manufacturer's instructions. In addition, the osteogenic differentiation potential of bone marrow-derived MSCs was assessed by Alizarin red S staining and alkaline phosphatase activity measurement. The inverted optical microscope was used to observe the culture medium (Nikon TE200, Japan).
MSCs were seeded into 4 wells of a 6-well plate at an initial density of 1×105 cells/well, and maintained in a humidified atmosphere of 5% CO2 at 37℃ for 24 h. Umbilical cord blood-derived CD34+ cells (1×105 cells/mL) were then plated onto the MSC layer in stem cell serum-free medium (Stem Cell Technologies, Vancouver, Canada); two wells were supplemented with 50 ng/mL of stem cell factor (SCF), Flt-3L (fms-like tyrosine kinase 3 Ligand), and thrombopoietin (TPO) (Stem Cell Technologies, Vancouver, Canada), and two wells were not. CD34+ HSCs (1×105) were also cultured in two other wells in stem cell serum-free medium supplemented with 50 ng/mL of SCF, Flt-3L, and TPO under the same conditions. All cultures were performed in a humidified atmosphere of 5% CO2 at 37℃ for 7 days. After 7 days of expansion, HSCs co-cultured with cytokines were separated into CD34+ and CD34− fractions using a small MACs column (Miltenyi Biotec, Bergisch Gladbach, Germany).
Total and viable cell numbers were counted at days 5 and 7 of culture using trypan blue stain 0.4% (Thermo Fisher Scientific, Waltham, MA, USA), and the stem cell percentage was evaluated by flow cytometry, according to the manufacturer's instructions (Partec, Gorlitz, Germany). Initially, 1×105 cells were stained with fluorescein isothiocyanate (FITC)-conjugated CD34 and phycoerythrin (PE)-conjugated CD38 at 4℃ for 30 mins. A replicate sample was stained with mouse IgG1 antibody (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA, Cat. N 11-4011-85, 12-4015-82) as an isotype control to ensure specificity.
In vitro colony-forming unit (CFU) assay was performed to detect the clonogenic potential of HSCs at days 0 and 7 of expansion using MethoCult H4434 methylcellulose medium (Stem Cell Technologies, Vancouver, Canada). After 14 days of culture, the number of colonies was counted.
Total RNA was extracted from fresh cord blood-derived CD34+ cells, following which the cells were expanded in all three culture conditions at days 0 and 7 of culture using TRIzol reagent, according to the manufacturer's instructions (Roche, Basel, Switzerland). The extracted RNA was immediately frozen and kept at −70℃ until use.
Reverse transcription was performed on 1 µg total RNA with AccuPower CycleScript RT PreMix Kit (Bioneer, Alameda, CA, USA). The reaction was carried out in 12 cycles of 22℃ for 30 s, 45℃ for 4 min, 55oC for 30 s, and 1 cycle of 95℃ for 5 min. Products were analyzed in 1.5% agarose gel under ultraviolet light. Quantitative RT-PCR was performed with the LightCycler system using 3 µL cDNA in 25 µL reaction volume, with 0.4 µM of each primer and 12.5 µL of 2X FastStart DNA Master SYBR Green I (Roche Molecular Biochemicals, Basel, Switzerland). The forward primer for DNMT1 was 5′-CCTCCAAAAACCCAGCCAAC-3′, the reverse primer was 5′-TCCAGGACCCTGGGGATTTC-3′, and the PCR product was 101 bp. Thermal cycling was initiated at 95℃ for 5 min and was followed by 40 cycles of PCR (95℃, 30 s; 57℃, 30 s; 72℃, 20 s). GAPDH was used as an endogenous control. Fold change ratio was calculated using Pfaffl method.
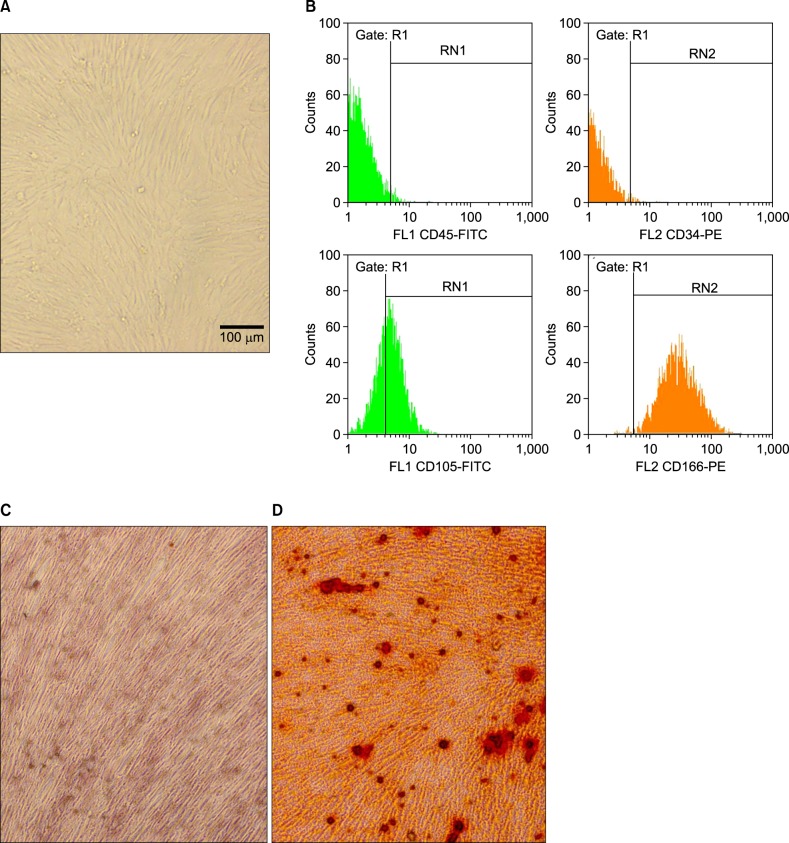
MSCs isolated from the bone marrow (Fig. 1A) were characterized by flow cytometric analysis of specific surface antigens. MSCs were found to be positive for the following adhesion molecules: CD44 (99.97%), CD166 (64%), CD73 (99.97%), CD29 (100%), CD105 (98.9%), and CD90 (90.92%), which are considered as markers for MSCs. The MSCs were negative for the hematopoietic lineage markers, CD34 and CD45 (Fig. 1B). The osteogenic differentiation potential of MSCs was evaluated to verify their efficiency. Both alizarin red staining and alkaline phosphatase activity results showed a positive reaction (Fig. 1C, D).
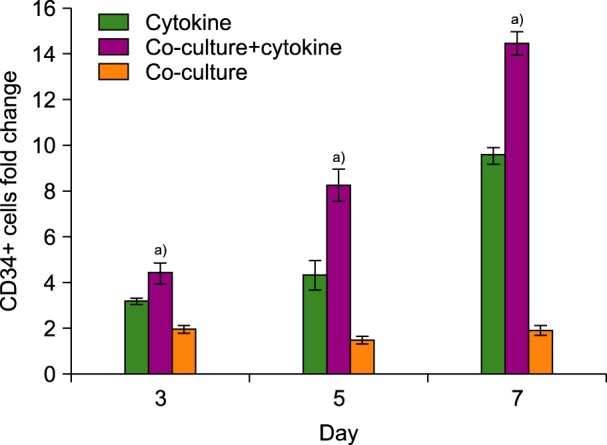
Purity of the separated CD34+ cells as determined by flow cytometry analysis was 86±12%. Moreover, 31.45±7% of them were positive for the CD38 marker, and the lineage-specific CD marker expression was low (N=3). Ex vivo expansion of human cord blood CD34+-enriched cells in serum-free medium supplemented with SCF, TPO, and Flt-3L was evaluated either with or without feeder layer (MSCs) using flow cytometry analysis. Fig. 2 shows the fold increase in CD34+ cells during 7 days of liquid cytokine culture, in the presence and absence of human MSCs. Maximum expansion was observed at day 7 of culture under all three conditions, during which the mean fold change of CD34+ cells was 9.5±4.6 in the cytokine-supplemented culture and 14.3±6.8 in the co-culture system with cytokines (N=6). However, in the co-culture system without cytokines, CD34+ cell numbers were increased up to 1.9±0.5. Therefore, the mean fold change of CD34+ cells at day 7 in the co-culture system with cytokines was significantly higher than in the cytokine culture without the MSCs feeder layer and in the co-culture system without cytokines (N=6, P<0.01).
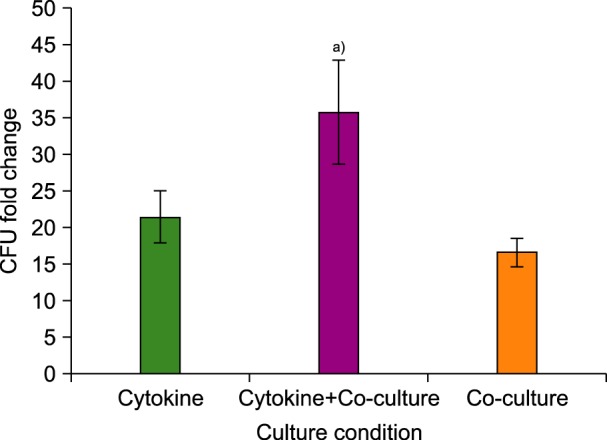
The highest CFU fold change was observed in the cytokine cultures with MSCs (co-culture) at day 7 (36±7.2). The CFU increase in cytokine cultures without MSCs at day 7 was 21.6±3.7 and in co-cultures with MSCs was 16.6±2. Therefore, the mean fold change of CFU at day 7 in both cytokine cultures, with and without MSCs feeder layer, was significantly higher than in the co-culture system without cytokines (N=3, P<0.01) (Fig. 3).
The expression level of DNMT1 in fresh CD34+ cells and in cells after 7 days of expansion in all three culture conditions was evaluated by RT-PCR. Results indicated that the fold change ratio of DNMT1 mRNA levels in the expanded cells (CD34+ cells expanded in co-culture system with cytokines, CD34− cells expanded in co-culture system with cytokines, cytokine cultures without MSCs feeder layer, and co-culture system without cytokines) and the fresh CD34+ cells was 2.1±0.4 (N=3), 0.5±0.2, 0.9±0.1, and 1.6±0.3 (N=3), respectively (Fig. 4). Thus, ex vivo expansion of CD34+ cells in both the co-culture systems resulted in a significant increase in DNMT1 expression levels. In addition, there was a significant difference between the expression level of DNMT1 in CD34+ and CD34− cells in the co-culture system with cytokines (N=3, P<0.01). However, there were no significant differences between the expression levels of DNMT1 in CD34+ cells in both the co-cultures conditions (with and without cytokines). Expression of DNMT1 in CD34+ cells in cytokine cultures without feeder layer was significantly lower than that in co-culture system with cytokines (N=3, P<0.05).
Expansion of CB-HSCs co-cultured with MSCs produces a more “natural” hematopoietic microenvironment for proliferation, self-renewal, and differentiation of CD34+ cells [15]. MSCs have been demonstrated to serve as a feeder layer and to maintain HSCs in an undifferentiated state [16]. In addition, DNMT1 plays an important role in self-renewal, niche retention, and differentiation of primitive HSCs [1217].
Here, mRNA expression levels of DNMT1 in CD34+ cells were compared in different culture conditions, with and without MSCs and cytokines during CB-HSC ex vivo expansion. Maximum expansion was observed at day 7 in the two cytokine-supplemented cultures, but in the co-culture system without cytokines, 2-fold expansion was constantly observed during the 7 days. Similar to our results, Alvarado-Moreno et al. (2010) have also previously reported that umbilical cord blood-derived CD34+ cells show significant proliferation after 14 days in cytokine cultures and maximum expansion is observed at day 7 of culture [17].
In this study, DNMT1 mRNA levels in the CD34+ cells expanded on MSCs feeder layer were increased compared to fresh CD34+ cells and there was no significant difference between the expression level of DNMT1 in CD34+ cells at both co-culture conditions, with and without cytokines. Expression of DNMT1 in CD34+ cells expanded in cytokine cultures without feeder layer was lower than in co-culture system with cytokines. Weidner et al. (2013) compared the DNA-methylation profiles of freshly isolated and culture-expanded HSCs either with or without MSCs. Their results demonstrated that all cultured HSCs, including those that remained CD34+, acquired significant DNA-hypermethylation; however, expansion had relatively little impact on DNA methylation, although proliferation was greatly increased by stromal support [18]. Therefore, our results suggest that HSC maintenance and proliferation during co-culture with MSCs might be mediated through an elevated expression of DNMT1. The molecular regulation of hematopoiesis is dependent on some epigenetic modifications, like DNA methylation. Epigenetic modifications play a critical role in the control of self-renewal and differentiation of HSCs. DNA methylation is an important epigenetic regulatory mechanism that regulates normal development by influencing gene transcription, genomic imprinting, and genome stability in mammalian cells [67]. DNMT1 regulates distinct patterns of methylation and expression of many critical genes in long-term HSCs and multipotent and lineage-restricted progenitors. It also plays a pivotal role in certain biological processes, including self-renewal, niche retention, and differentiation of primitive HSCs. However, the precise molecular mechanism through which DNMT1 is involved in HSCs maintenance remains unknown [1011].
In conclusion, based on the above-mentioned evidences, an elevated expression of DNMT1 has been shown to be associated with increased expansion and proliferation of CD34+ cells co-cultured with MSCs as a feeder layer. Therefore, DNMT1 may be considered as a potential factor in the maintenance of expanded HSCs co-cultured with human MSCs. However, further studies are required to identify the exact mechanism by which MSCs regulate HSC differentiation and other biological processes.
References
1. Delaney C, Ratajczak MZ, Laughlin MJ. Strategies to enhance umbilical cord blood stem cell engraftment in adult patients. Expert Rev Hematol. 2010; 3:273–283. PMID: 20835351.

2. Broxmeyer HE. Umbilical cord transplantation: epilogue. Semin Hematol. 2010; 47:97–103. PMID: 20109617.

3. Dahlberg A, Delaney C, Bernstein ID. Ex vivo expansion of human hematopoietic stem and progenitor cells. Blood. 2011; 117:6083–6090. PMID: 21436068.

4. Conrad PD, Emerson SG. Ex vivo expansion of hematopoietic cells from umbilical cord blood for clinical transplantation. J Leukoc Biol. 1998; 64:147–155. PMID: 9715252.

5. Alenzi FQ, Alenazi BQ, Ahmad SY, Salem ML, Al-Jabri AA, Wyse RK. The haemopoietic stem cell: between apoptosis and self renewal. Yale J Biol Med. 2009; 82:7–18. PMID: 19325941.
6. Wang T, Nandakumar V, Jiang XX, et al. The control of hematopoietic stem cell maintenance, self-renewal, and differentiation by Mysm1-mediated epigenetic regulation. Blood. 2013; 122:2812–2822. PMID: 24014243.

7. Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008; 454:766–770. PMID: 18600261.

8. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998; 19:219–220. PMID: 9662389.

9. Lei H, Oh SP, Okano M, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996; 122:3195–3205. PMID: 8898232.

10. Sashida G, Iwama A. Epigenetic regulation of hematopoiesis. Int J Hematol. 2012; 96:405–412. PMID: 23054647.

11. Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009; 5:442–449. PMID: 19796624.

12. Bröske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009; 41:1207–1215. PMID: 19801979.

13. Majumdar MK, Thiede MA, Haynesworth SE, Bruder SP, Gerson SL. Human marrow-derived mesenchymal stem cells (MSCs) express hematopoietic cytokines and support long-term hematopoiesis when differentiated toward stromal and osteogenic lineages. J Hematother Stem Cell Res. 2000; 9:841–848. PMID: 11177595.

14. Mehrasa R, Vaziri H, Oodi A, et al. Mesenchymal stem cells as a feeder layer can prevent apoptosis of expanded hematopoietic stem cells derived from cord blood. Int J Mol Cell Med. 2014; 3:1–10. PMID: 24551815.
15. Yildirim S, Boehmler AM, Kanz L, Mohle R. Expansion of cord blood CD34+ hematopoietic progenitor cells in coculture with autologous umbilical vein endothelial cells (HUVEC) is superior to cytokine-supplemented liquid culture. Bone Marrow Transplant. 2005; 36:71–79. PMID: 15895114.

16. Wagner W, Saffrich R, Ho AD. The stromal activity of mesenchymal stromal cells. Transfus Med Hemother. 2008; 35:185–193. PMID: 21547116.

17. Alvarado-Moreno A, Chávez-Gonzalez A, Cérbulo A, Arriaga L, Mayani H. In vitro cell cycle dynamics of primitive hematopoietic cells from human umbilical cord blood. Hematology. 2010; 15:11–20. PMID: 20132657.
18. Weidner CI, Walenda T, Lin Q, et al. Hematopoietic stem and progenitor cells acquire distinct DNA-hypermethylation during in vitro culture. Sci Rep. 2013; 3:3372. PMID: 24284763.

Fig. 1
(A) MSCs isolated from the bone marrow (×20). (B) Characterization of MSCs using flow cytometry. Evaluation of osteogenic differentiation of MSCs cultured under control conditions (C) and osteogenic conditions (D).




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download