INTRODUCTION
Up to 70% of acute myeloid leukemia (AML) patients achieve remission after receiving the most commonly used induction regimen, which is a combination of cytarabine and anthracycline [
1]. However, the majority of AML patients who achieve complete remission (CR) experience recurrence within 3 years after diagnosis [
2]. AML relapse is usually associated with a dismal prognosis. No established standard salvage regimen that definitely prolongs survival has been approved, although there have been various attempts at improving the response to re-induction chemotherapy, including high-dose cytosine arabinoside (Ara-C) and fludarabine. A range of biologically targeted therapies has been developed for the treatment of relapsed AML [
3].
Early relapse (duration of remission prior to relapse, <6 mo), adverse cytogenetics at initial diagnosis, older age at the time of relapse, and prior hematopoietic stem cell transplantation (HCT) are poor prognostic factors in relapsed AML [
2]. The cytogenetic risk category at diagnosis is the single most important independent prognostic factor in relapsed AML as well as in newly diagnosed AML [
4]. It is imperative that we predict the prognosis at the time of relapse, because only a few patients with relapsed AML experience durable clinical benefits from current re-induction therapy [
5].
Some studies have analyzed the cytogenetics at diagnosis and relapse in relapsed AML patients. The frequencies of karyotype change between the 2 time points have been reported as 28–52% [
678]. Whole-genome sequencing has shown that AML relapse is associated with the occurrence of new mutations and clonal evolution [
9]. A German study analyzed cytogenetic findings in 117 patients with AML at both diagnosis and relapse. The authors reported that the interval between diagnosis and relapse was significantly shorter in cases with an evolution of the aberrant karyotype or an unrelated karyotype at relapse, compared to cases with no alterations or with only the regression of the aberrant karyotype at relapse (9.2±4.4 vs. 14.0±8.5 mo, respectively;
P =0.0081) [
10]. A recent Korean study reported that karyotype differences between diagnosis and relapse were significantly associated with shorter overall survival (OS) (
P =0.023; RR=2.655) and event-free survival (EFS) (
P =0.033; RR=2.831) [
11].
Thus far, conclusive data regarding the utility of reassessing the cytogenetic aberration at relapse have not been reported. In the present study, we reviewed 45 patients with relapsed AML, who had been treated at a single institution.
Go to :

MATERIALS AND METHODS
We reviewed the medical records and laboratory findings of AML patients who had relapsed between 2004 and 2013, and had been treated at the Division of Hematology of Pusan National University Hospital. Four patients with AML of the French-American-British classification M3 with t(15;17) (q24;q21) were excluded from the present study and 55 patients with relapsed AML were included. The diagnosis and classification were established by bone marrow (BM) aspiration with biopsy. Written informed consent for genetic test and research were obtained from all patients at the time of each bone marrow biopsy.
Cytogenetic abnormality analysis of the BM aspirate was conducted using the standard banding technique. The results of fluorescence in situ hybridization (FISH) and reverse transcriptase (RT)-polymerase chain reaction (PCR) were available for 44 out of 55 patients at the time of diagnosis whereas results of FISH and RT-PCR analyses were available for only 16 out of 55 patients at the time of relapse. Of the 55 patients, the results of karyotype analysis at the time of diagnosis and relapse were available for 51 patients and 42 patients, respectively. Karyotype records were not available for patients who were not karyotyped or in the absence of mitosis. Risk category classification was conducted using the new European Leukemia Net recommendations [
2].
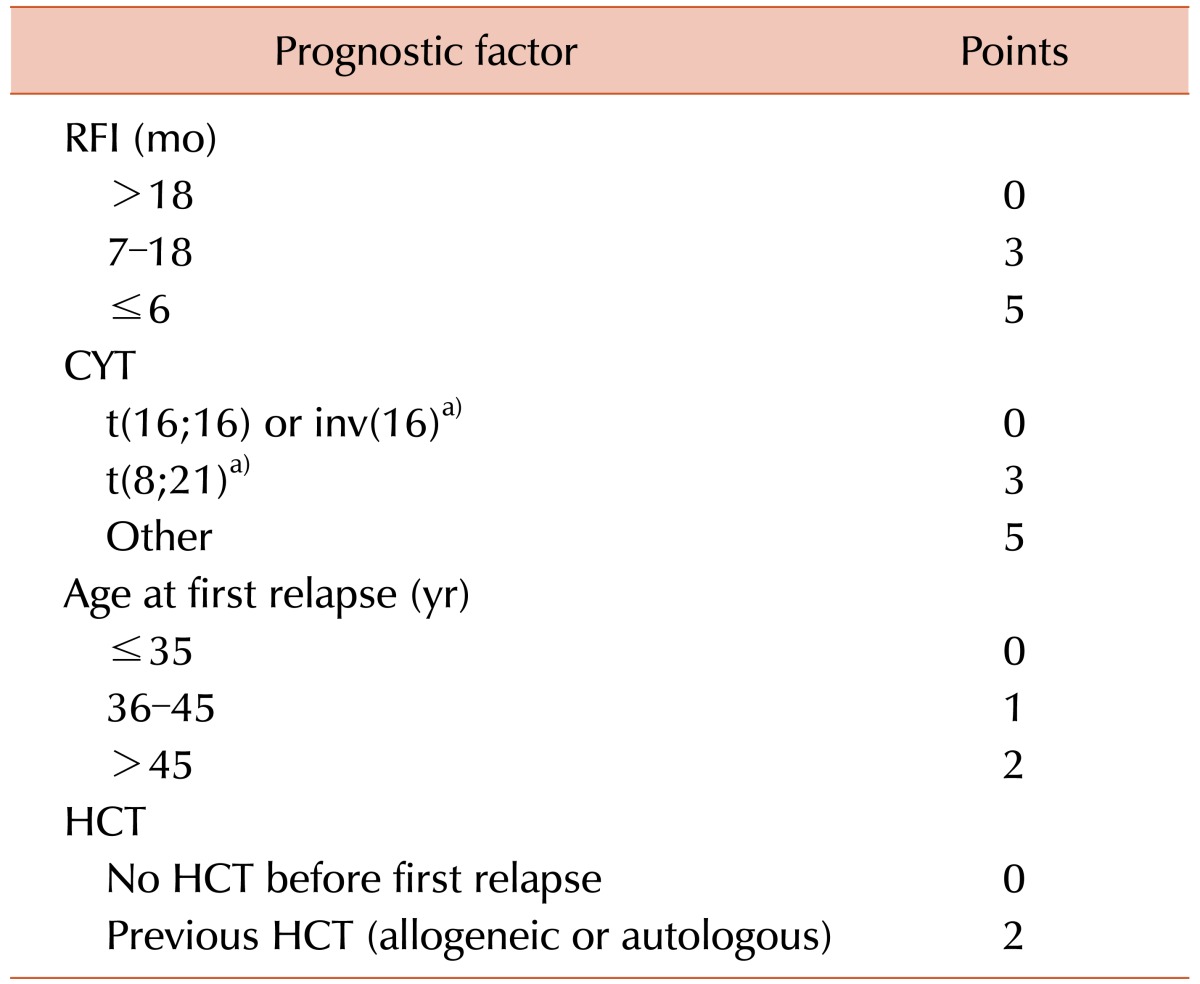
Based on a multivariate analysis of 667 AML patients at first relapse, Breems et al. [
5] reported the prognostic index for adult patients with AML. We studied 55 patients based on this scoring system (
Table 1).
Table 1
Prognostic score for AML at first relapse.

CR was defined by a normal cellular BM with <5% myeloblasts and normalization of the peripheral blood counts (neutrophils>1×10
9/L and platelets>100×10
9/L). Relapse was defined by the detection of at least 5% leukemic blasts in the BM aspirates of patients with previously documented CR [
12]. Relapse-free interval (RFI) refers to the interval between the documented dates of CR and relapse. OS refers to the period between the dates of diagnosis and death. EFS and post-relapse survival (PRS) were defined as the periods from the date of diagnosis to relapse and from the date of relapse to death, respectively. All statistical analyses were performed using the SPSS software version 18 (SPSS, Inc., Chicago, IL, USA). All
P values <0.05 were considered statistically significant.
Ethics statement
The present study was approved by the institutional review board of the Pusan National University Hospital in Busan, Korea. Informed consent for medical record analysis was waived by the board.
Go to :

RESULTS
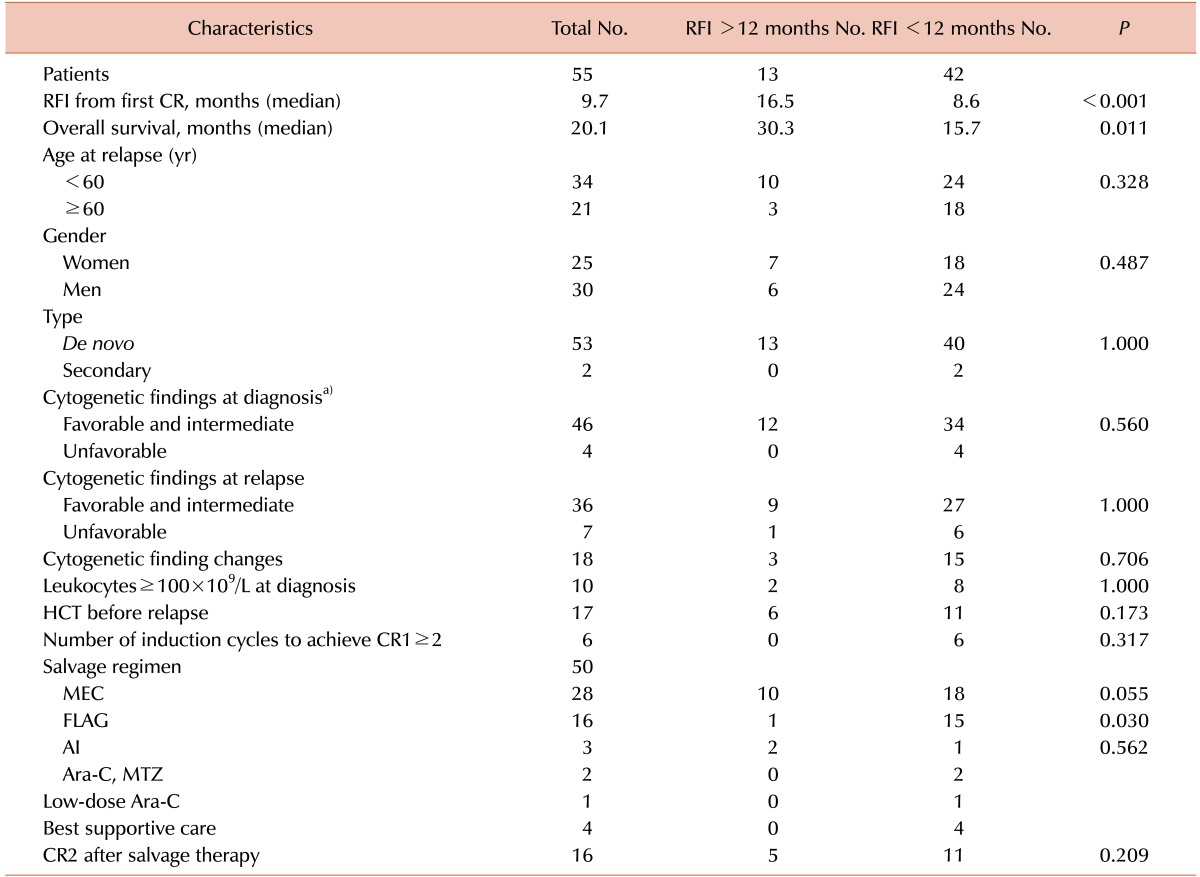
The median OS and RFI durations for the 55 relapsed AML patients with a median age of 57 years at the time of relapse were 20.1 and 9.7 months, respectively (
Table 2). There were two secondary AML patients. One of the patients was treated with 6 cycles of cyclophosphamide, Adriamycin, vincristine, and prednisone (CHOP) owing to peripheral T cell lymphoma and showed metabolic CR while the other patient was treated with concurrent chemoradiation using 6 cycles of paclitaxel and carboplatin owing to cervical cancer.
Table 2
Patient characteristics.

The results of cytogenetic analyses were not available for 4 patients at the time of diagnosis because they were under observation, lacked mitosis, or the initial data from another hospital were unavailable. The cytogenetic findings were sorted according to the European Leukemia Net recommendations. The karyotypes for 42 out of 55 patients (76%) were available at both diagnosis and relapse. Cytogenetic alterations were observed between diagnosis and relapse for 18 out of 42 patients (43%).
Induction therapy with idarubicin (12 mg/m2/d, intravenous [IV] for 15 min on days 1–3) and Ara-C (100 mg/m2/d, continuous IV infusion on days 1–7; AI73) was administered to all patients except for one 74-year-old patient, who received daunorubicin (45 mg/m2/d, IV for 15 min on days 1-3) and Ara-C (100 mg/m2/d, continuous IV infusion on days 1–7).
A second CR was achieved in 3 out of 51 patients who were treated with the fludarabine, Ara-C, and granulocyte colony-stimulating factor (G-CSF) regimen (FLAG; fludarabine, 30 mg/m2/d IV for 2 hours on days 1–5; G-CSF, 300 µg subcutaneous injection daily and 12 hours before fludarabine on days 0–5; and Ara-C, 2.0 g/m2/d IV for 4 hours and 4 hours after fludarabine on days 1–5), and received allogeneic hematopoietic stem cell transplantation (HCT) as a consolidation therapy. The most frequently used salvage regimen was mitoxantrone, etoposide, and cytarabine (MEC; mitoxantrone, 12 mg/m2/d on days 1–4; etoposide, 100 mg/m2/d on days 5–7; and Ara-C, 1.5 g/m2/12 hours on days 1–4) in 28 patients.
Univariate analysis with the log-rank test showed that OS was associated with an unfavorable karyotype at the time of diagnosis (unfavorable vs. favorable or intermediate karyotype at diagnosis, 8.5 vs. 21.8 mo,
P =0.001;
Fig. 1), salvage treatment (salvage treatment done vs. not done, 21.3 vs. 12.9 mo,
P =0.002), and RFI>12 months (RFI>12 vs. ≤12 months, 30.3 vs. 15.7 mo,
P =0.011). The log-rank test showed that karyotype changes and unfavorable karyotype at relapse did not show significant correlations with OS (changed vs. unchanged karyotype, 14.4 vs. 21.8 mo,
P =0.927; unfavorable vs. favorable or intermediate karyotype at relapse, 8.5 vs. 21.2 mo,
P =0.136).
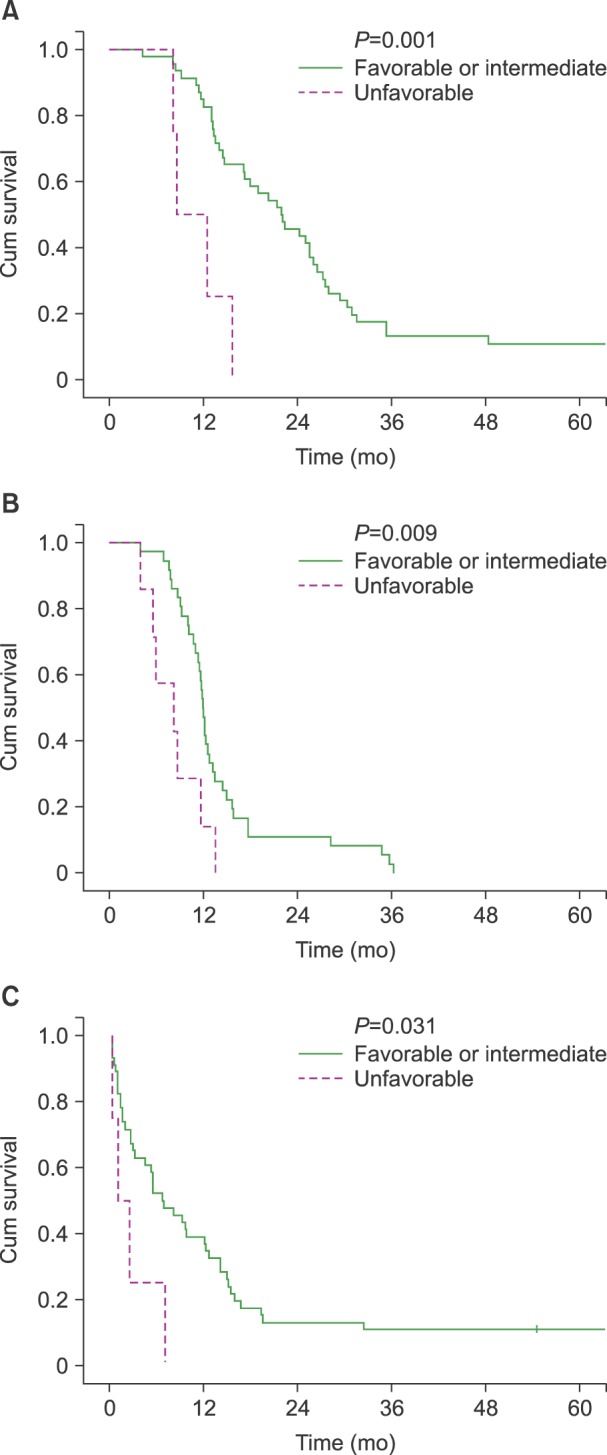 | Fig. 1(A) Overall survival (OS) of acute myeloid leukemia (AML) patients according to the cytogenetic group at diagnosis. (B) Event-free survival (EFS) in AML patients according to the cytogenetic group at relapse. (C) Post-relapse survival (PRS) in AML patients according to the cytogenetic group at diagnosis.
|
The variables included in the multivariate analyses using the Cox proportional hazards model were unfavorable karyotype at diagnosis, salvage treatment, the RFI, and 2 known factors, which are the age at relapse and HCT before relapse. The Cox proportional hazards model showed that an unfavorable karyotype at diagnosis (hazard ratio [HR]=6.327, P =0.002), salvage treatment (HR=0.223, P =0.012), and RFI (HR=0.872, P =0.001) were important factors associated with the OS. The RFI from the first CR could be interpreted as a 1 month longer RFI related to the 0.872 times decreased HR for OS (95% confidence interval=0.804–0.946). According to the Cox proportional hazards model, the salvage FLAG treatment was a significant factor for OS (HR=0.399, P =0.010). However, in the same model, the salvage MEC treatment did not show reduced HR for OS.
The log-rank test showed that EFS was shorter in patients with unfavorable karyotype subsets than in patients with favorable or intermediate subsets at both diagnosis and relapse (unfavorable vs. favorable or intermediate karyotypes at diagnosis, 8.2 vs. 11.9 mo,
P =0.003; unfavorable vs. favorable or intermediate karyotypes at relapse, 8.2 vs. 11.9 mo
P =0.009;
Fig. 1). Karyotype change was not a significant factor for EFS (changed vs. unchanged karyotype, 11.3 vs. 11.9 mo,
P =0.185).
Univariate analysis identified unfavorable karyotype at diagnosis (1.0 vs. 6.7 mo,
P =0.031), salvage treatment (6.8 vs. 0.7 mo,
P =0.001), and RFI >6 months (8.1 vs. 2.7 mo,
P =0.019) as significant factors for PRS (
Fig. 1). Karyotype changes were not significantly associated with PRS (2.7 vs. 6.8 mo,
P =0.748). Multivariate analysis using Cox model adjusted for karyotype at diagnosis, salvage treatment, RFI (significant in log-rank tests), age at relapse, and HCT before relapse revealed that salvage FLAG was significantly beneficial (HR=0.447,
P =0.031) whereas salvage MEC was not (HR=1.801,
P =0.105).
We categorized the 55 patients using the scoring system by Breems et al. [
5], which is based on the prognostic index for adult patients with AML at first relapse (
Table 1). The favorable and intermediate risk groups had similar survival curves, while the poor risk group had a distinctly declining survival curve (
Fig. 2).
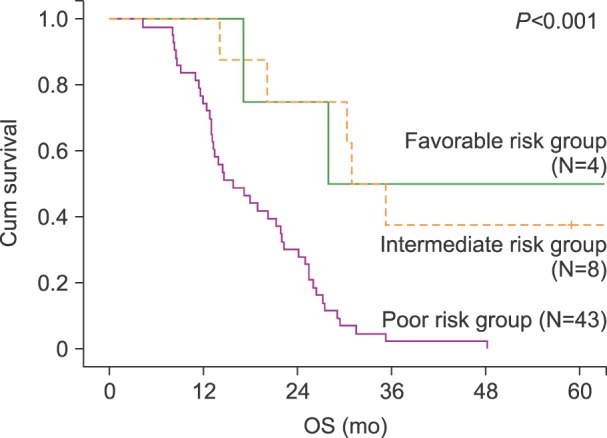 | Fig. 2Overall survival (OS) according to prognostic scoring for acute myeloid leukemia (AML) at first relapse. The scores for patients in the favorable risk group were 1–6 points, those for patients in the intermediate risk group were 7–9 points, and those for the patients in the poor risk group were 10–14 points.
|
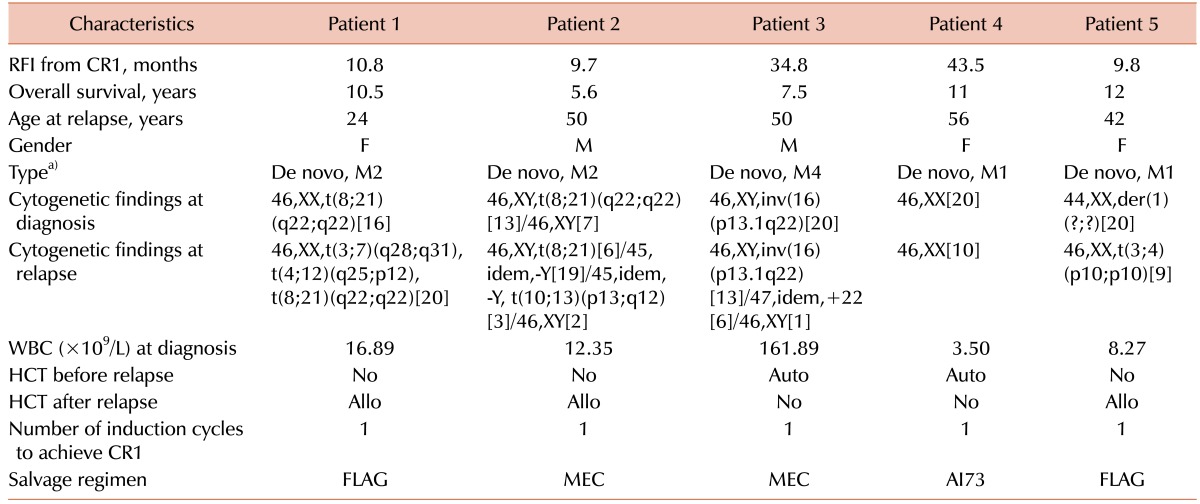
Five patients survived longer than 5 years (
Table 3); all 5 patients had achieved CR after 1 cycle of AI73. We treated patient 3 with high-dose Ara-C and patient 4 with idarubicin (12 mg/m
2/d, IV for 15 min on days 1–2) and Ara-C (100 mg/m
2/d, IV continuous infusion on days 1–5; AI52) as consolidation therapies. These 5 patients were followed up without any sign of recurrence at the last follow-up date (Sep. 2015–Jan. 2016).
Table 3
Patients who were alive at the last follow-up.

Go to :

DISCUSSION
In the present study, karyotype changes were observed in 18 out of 42 patients (43%) with karyotypes available at both diagnosis and relapse. This finding is consistent with the 38% (44/117 and 17/45) reported by other recent studies on karyotype changes in relapsed AML [
1011]. Older studies reported variable changes in the karyotypes of patients between diagnosis and relapse (61% [63/103], 52% [87/168], and 28% [17/60]) [
678]. Altogether, most studies including the present one found that karyotype changes were identified in <50% of relapsed AML patients. This finding needs further investigation to identify whether missing karyotype analysis results at relapse would change the conclusion or whether genetic analysis would be required.
Karyotypic change has been suggested as a poor prognostic factor of relapsed acute lymphoblastic leukemia [
1314]. Although we could not establish a significant association between karyotypic changes and OS (14.4 vs. 21.8 mo,
P =0.927), EFS (11.3 vs. 11.9 mo,
P =0.185), and PRS (2.7 vs. 6.8 mo,
P =0.748), we still cannot rule out a possible correlation between them. The absence of statistically significant karyotypic changes does not preclude the clinical relevance of karyotype analysis in patients with relapsed AML at the time of relapse. The median number of non-synonymous somatic mutations for AML is believed to be approximately 8 [
15]. Although this is far less than that for solid tumors, the genetic heterogeneity among cancer cells can affect the response to therapy. Because it can provide potentially valuable information, genetic analysis should routinely be performed at the time of relapse and its use should be established through further investigation.
The patients who received salvage treatment had longer OS and PRS (salvage treatment done vs. not done, 21.3 vs. 12.9 mo,
P =0.002, 6.8 vs. 0.7 mo,
P =0.001, respectively). The Cox proportional hazards models for OS and PRS were adjusted for this factor. Multivariate analysis using the Cox proportional hazards model showed that salvage FLAG treatment was particularly related to a longer OS compared to non-FLAG salvage treatment (HR=0.399,
P =0.010). However, the non-FLAG group was heterogeneous, which is a limitation in terms of establishing a reliable conclusion. Non-FLAG salvage treatment includes all regimens of MEC, AI, Ara-C/mitoxantrone, low-dose Ara-C, and best supportive care (
Table 2).
The FLAG regimen is generally considered intensive and toxic. If residual disease has a higher influence on the progress of relapsed AML patients than clonal evolution, the administration of an intensive salvage regimen could be reasonable. Recently, a team at the Washington University School of Medicine reported that the detection of persistent leukemia-associated mutations in BM cells in day 30 remission samples was associated with a significantly increased risk of relapse and reduced OS [
16]. In the present study, we used whole-genome or exome sequencing to determine whether genomic approaches can provide novel prognostic information, especially in patients with intermediate risk AML. While the analysis of comprehensive genomic data from 71 patients did not improve the outcome assessment over current standard-of-care metrics, 24 patients with persistent leukemia-associated mutations in at least 5% of BM cells at remission had significantly reduced EFS and OS. This suggests that residual disease might play a more important role than clonal evolution, which is considered a major cause of resistance against therapy. Further studies aimed at establishing the superiority of the FLAG regimen as a salvage treatment could be valuable.
The present study had some limitations. The FISH and RT-PCR data on molecules including FLT3 or NPM1 were available for only 12 patients at diagnosis and relapse. Moreover, a different list of molecular data was available for each patient and time point. There were 3 patients with changes between the molecular data at diagnosis and relapse; however, the changes did not affect survival significantly. Therefore, our analysis included information on karyotypes and various other factors that we were able to obtain. Another limitation is that the present study was a retrospective one conducted using a small number of patients.
In conclusion, the salvage FLAG regimen could be an affordable treatment option for relapsed AML. In order to improve the survival of relapsed AML patients, the prognostic factors affecting survival should be established clearly and more sophisticated molecular studies should be conducted.
Go to :









 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download