Abstract
Background
The C-X-C chemokine receptor 7 (CXCR7) has been shown to be a decoy receptor for CXCR4 in certain cell types. We investigated the expression status and functional roles of CXCR7 in acute myeloid leukemia (AML) cells in vitro.
Methods
CXCR7 mRNA was knocked down in AML cells by using small interfering RNA (siRNA) technology, and subsequent biological alterations in the cells were evaluated in vitro.
Results
All AML cell lines examined in this study (U937, K562, KG1a, HL-60, and MO7e) and primary CD34+ cells obtained from patients with AML expressed CXCR7 mRNA at various levels. Western blotting showed that all AML cells produced CXCR7. Furthermore, all AML cells expressed CXCR7 in both the cytoplasm and on the cell surface at various levels. Stromal cell-derived factor-1 (SDF-1; C-X-C motif ligand 12 (CXCL12)) induced internalization of cell surface CXCR7. However, neither hypoxia nor the examined hematopoietic growth factors (interleukin-1β (IL-1β), IL-3, IL-6, granulocyte-colony-stimulating factor, granulocyte, macrophage-colony-stimulating factor, and stem cell factor) and proinflammatory cytokines (interferon-γ, transforming growth factor-β, and tumor necrosis factor-α) were found to alter cell surface CXCR7 expression. The transfection of AML cells with CXCR4 siRNA, but not CXCR7 siRNA, significantly impaired the CXCL12-induced transmigration of the cells. The transfection of AML cells with CXCR7 siRNA did not affect the survival or proliferation of these cells. Knockdown of CXCR7, but not CXCR4, induced the upregulation of CXCL12 mRNA expression and CXCL12 production in AML cells.
Stromal cell-derived factor 1 (SDF-1), also known as C-X-C motif ligand 12 (CXCL12), is a chemokine that is constitutively expressed and produced by bone marrow stromal cells (BMSCs). It induces the migration and homing of hematopoietic stem cells (HSCs) and progenitor cells (HPCs) by signaling via the G protein-coupled receptor, C-X-C chemokine receptor 4 (CXCR4) [1]. Acute myeloid leukemia (AML) cells also express CXCR4 and respond to CXCL12 [23], resulting in the trafficking of these cells to the bone marrow (BM) [34]. CXCL12 alone has negligible effects on the proliferation of normal and malignant hematopoietic cells in vitro [5], but the CXCL12/CXCR4 axis has been demonstrated to be involved in the development and progression of AML [46].
CXCR4 was the only known receptor for CXCL12 until the orphan receptor RDC1 (later renamed CXCR7) was discovered as an additional receptor for this chemokine [7]. Thereafter, CXCR7-deficient mice revealed cardiovascular defects and early postnatal lethality, but normal hematopoiesis [8]. Despite the controversial roles of CXCR7, especially with respect to CXCR4, it has been shown that CXCR7 is expressed in many cancer cell types and is involved in the development and progression of various cancers [9101112]. Additionally, overexpression of CXCR7 in cancer cells was shown to indicate poor prognosis in many types of cancers [131415]. Based on these observations, it has been proposed that CXCR7 may be a therapeutic target in some cancers.
Whereas CXCR7 protein is expressed by primitive red blood cells (RBCs) during murine embryonic development, in adult mammals, this protein is not expressed by normal peripheral blood (PB) cells [16]. A recent study has shown that CXCR7 is expressed at very low levels on normal CD34+ human HPCs and does not play a direct role in their proliferation or migration; however, it is involved in the trafficking/ adhesion of human leukemic cells [17].
The roles of CXCR7 in the survival and growth of AML cells, however, are not fully understood. Thus, the aim of the present study was to determine the role(s) of CXCR7 expression in the survival and proliferation of AML cells. Using siRNA technology, CXCR7 was knocked down in AML cells and subsequent biological alterations occurring in the cells were evaluated in vitro.
BM samples were obtained, with informed consent, from 5 patients with AML at the time of diagnosis. CD34+ cells were purified from PB using the MACS system (Miltenyi Biotec, Auburn, CA, USA). Only cell preparations that contained >95% CD34+, as assessed by flow cytometry, were used in the experiments.
The human AML lines U937, HL-60, K562, and KG1a were purchased from the American Type Culture Collection (Manassas, VA, USA). The U937 cells were cultured in RPMI-1640 medium (Gibco-BRL Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL Life Technologies, Grand Island, NY, USA). The K562, KG1a, and HL-60 cells were grown in Iscove's modified Dulbecco's medium (IMDM; Gibco-BRL Life Technologies, Grand Island, NY, USA) supplemented with 10% FBS. The MO7e cells were grown in IMDM supplemented with 10% FBS and 10 ng/mL granulocyte macrophage colony-stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN, USA). For hypoxic exposure, cells were incubated with 5% CO2 and 1% O2 (v/v), balanced with N2 gas, at 37℃ for the time-periods indicated. The recombinant human CXCL12 and cytokines examined in this study were purchased from R& Systems. The following cytokines were used: interleukin-6 (IL-6), IL-3, granulocyte colony-stimulating factor (G-CSF), stem cell factor (SCF), thrombopoietin (TPO), transforming growth factor-beta (TGF-β), tumor necrosis factor-alpha (TNF-α), and interferon-gamma (IFN-γ).
The cells were incubated with fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, or allophycocyanin (APC)-conjugated monoclonal antibodies at 4℃ for 30 min and analyzed using a Coulter Elite flow cytometer (Coulter Electronics Ltd., Hialeah, FL, USA) or a FACSCanto II flow cytometer (BD Pharmingen, San Diego, CA, USA). The monoclonal antibodies used were FITC-conjugated anti-CXCR4, PE-conjugated anti-CXCR4 (clone 12G5; BD Pharmingen, San Diego, CA, USA), and APC-conjugated anti-CXCR7/RDC-1 (clone 11G8; R&D Systems, Minneapolis, MN, USA). To detect cytoplasmic CXCR4 or CXCR7 expression, the cells were permeabilized with a saponin-based reagent (BD Pharmingen, San Diego, CA, USA) and labeled. To detect apoptosis, cells were stained with FITC-conjugated Annexin V and analyzed by flow cytometry.
Cells grown on coverslips (Paul Marienfeld Gmbh & Co. KG, Lauda-Koenigshofen, Germany) were washed with cold phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde (Sigma-Aldrich Co., Saint Louis, MO, USA) for 15 min at 37℃, and washed 3 times with PBS. The cells were then incubated with a murine monoclonal anti-CXCR4 antibody (1:2,000; clone 12G5; R&D Systems, Minneapolis, MN, USA) and a murine monoclonal anti-CXCR7/RDC-1 antibody (1:2,000; clone 11G8; R&D Systems, Minneapolis, MN, USA) for 90 min at 37℃, washed 3 times with PBS, and incubated with FITC- or PE-conjugated anti-mouse IgG (1:4,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at 37℃ for 60 min. The cells were washed 3 times with PBS, fixed, mounted on glass slides with PBS, and observed under a laser scanning confocal microscope (Olympus Corp., Lake Success, NY, USA).
For the transmigration experiments, cells (2×105 cells/well) were loaded into the upper chamber of a 24-well Transwell plate containing a 5 µm microporous membrane (Corning-Costar, Tewksbury, MA, USA), and the cells were allowed to migrate into the lower chamber containing 200 ng/mL CXCL12 for 4 hr. The migrated cells were counted, and the percentage of migrated cells or fold-increase in the number of migrated cells relative to that of the control (i.e., the migration index) was calculated.
Cell proliferation was measured using a colorimetric assay kit (CCK-8 assay kit; Dojindo Laboratories, Tokyo, Japan) according to the manufacturer's instructions. Briefly, 5×103 cells were incubated in 96-well plates in serum-free X-VIVO medium (BioWhittaker, Walkersville, MA, USA). After incubation, 10 µL CCK-8 solution, provided by the manufacturer, was added to each well. The optical density (OD) was measured 3 hr later using a spectrophotometer (Molecular Devices Co., Sunnyvale, CA, USA). The proliferation index represents the fold increase in the OD of the experimental cell solution from that of the control. The relative proliferation index represents the fold increase in the OD of the experimental cell solution from that of the control at the beginning of the incubation.
Total RNA was prepared from cells using TRIzol reagent (Gibco-BRL Life Technologies, Grand Island, NY, USA), according to the manufacturer's instructions. After purification, 1 µg RNA was reverse-transcribed using SuperScript reverse transcriptase (Gibco-BRL Life Technologies, Grand Island, NY, USA) and the universal primer oligo (dT)15 (Promega, Madison, WI, USA). In each reaction, 1 µL of cDNA was added to 24 µL PCR buffer (Gibco-BRL Life Technologies, Grand Island, NY, USA) supplemented with 2 mM MgCl2, 0.2 µM of each primer, and 1-U Koma Taq polymerase (Koma International, Seoul, Korea). Using a GeneAmp PCR system (Perkin Elmer, Norwalk, CT, USA), 30 cycles of 1 min at 94℃, 45 sec at 55-65℃, and 1 min at 72℃ were performed. The following primers were used: human CXCL12 (sense, AGA ATT CAT GAA CGC CAA GG; anti-sense, AGG ATC CTC ACA TCT TGA ACC); human CXCR4 (sense, AAT CTT CCT GCC CAC CAT CTA CTC C; antisense, GCG GTC ACA GAT ATA TCT GTC ATC TGC C); human CXCR7 (sense, ACG TGG TGG TCT TCC TTG TC; antisense, AAG GCC TTC ATC AGC TCG TA); and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sense, CAT GTG GGC CAT GAG GTC CAC CAC; antisense, TGA AGG TCG GAG TCA ACG GAT TTG GTC).
The quantification of human CXCL12 and GAPDH mRNA was performed by 2-step RQ-PCR on a Rotor-Gene 6000 thermal cycler (Corbett Research, Mortlake, Victoria, Australia) using SyBR Green PCR Master Mix reagent (Qiagen, Hilden, Germany). The amplification conditions were as follows: 15 min at 95℃ for activation, 50 cycles at 95℃ for 10 sec for denaturation, annealing at 50-65℃ for 15 sec, and extension at 72℃ for 20 sec. The following primers were used: human CXCR4 (sense, AAT CTT CCT GCC CAC CAT CTA CTC C; antisense, GCG GTC ACA GAT ATA TCT GTC ATC TGC C); human CXCR7 (sense, ACG TGG TGG TCT TCC TTG TC; antisense, AAG GCC TTC ATC AGC TCG TA); human CXCL12 (sense, AGA ATT CAT GAA CGC CAA GG; anti-sense, AGG ATC CTC ACA TCT TGA ACC); and human GAPDH (sense, CAT GTG GGC CAT GAG GTC CAC CAC; antisense, TGA AGG TCG GAG TCA ACG GAT TTG GTC).
Western blotting was used to detect CXCL12. Cells were starved in serum-free medium for 12 hr and then collected by centrifugation, washed in PBS, and lysed using sodium dodecyl sulfate (SDS) sample buffer (187.5 mM Tris-HCl, pH 6.8, 6% (w/v) SDS, 100% glycerol, 150 mM dithiothreitol, and 0.03% (w/v) bromophenol blue). Equal amounts of protein from each sample were separated by electrophoresis on 10% SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Amersham Life Science, Arlington Heights, IL, USA). The membranes were blocked for 1 hr in Tris-buffered saline (TBS) containing 5% (w/v) milk and 0.1% Tween 20 and then incubated with a primary mouse monoclonal antibody (Cell Signaling Technology Inc., Danvers, MA, USA) overnight at 4℃. The blots were washed with TBS containing Tween 20, incubated with the secondary antibody for 2 hr, and developed using West-Zol Plus (iNtRON Biotechnology, Seoul, Korea). Anti-mouse CXCL12 polyclonal antibody (Thermo Scientific, Barrington, IL, USA) was used.
Cells were seeded in 12-well plates (3×105 cells/well), incubated for 5-10 min at room temperature, and then transfected with 5-25 nM CXCL12 siRNA or control siRNA (Qiagen, Hilden, Germany) using the HiperFect transfection reagent (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Cells were cultured in normal growth medium for 24 hr after the transfection. The transfection efficiency of the control siRNA was evaluated by fluorescence microscopy. The suppression of CXCR4 and CXCR7 mRNA expression levels was determined by RQ-PCR.
All AML cell lines examined in this study (U937, K562, KG1a, HL-60, and MO7e) expressed both CXCR4 and CXCR7 mRNA at various levels, as shown by semi-quantitative and RT-PCR analysis. This expression was confirmed by RQ-PCR (data not shown). Western blotting showed that all of the AML cell lines produced CXCR7 protein. In parallel, all the AML cell lines, as well as the primary CD34+ cells obtained from the BM of patients with AML, expressed CXCR7 on the cell surface at various levels, as revealed by flow cytometric analysis. Flow cytometry after cell permeabilization revealed that abundant CXCR4 and CXCR7 were present in the cytoplasm of these cells lines. The presence of CXCR7 on the cell surface, as well as in the cytoplasm in U937 cells, was confirmed by immunofluorescence staining (Fig. 1).
CXCL12 induced the internalization of cell surface CXCR7 in the U937 cells (Fig. 2). Subsequently, we examined whether certain cytokines affected the expression of CXCR7 in AML cells. The AML cell lines were incubated with cytokines at various concentrations for up to 72 hr and then CXCR7 expression on the cell surface was examined using flow cytometry. The hematopoietic growth factors (IL-1β, IL-3, IL-6, G-CSF, GM-CSF, and SCF) and proinflammatory cytokines (IFN-γ, TGF-β, and TNF-α) examined did not alter the cell surface CXCR7 expression on the U937 cells. Similar results were obtained in the K562, KG1a, HL-60, and MO7e cells (data not shown). Exposure of the U937 cells to hypoxia (5% CO2 and 1% O2, balanced with N2 gas) at 37℃ for up to 12 hr did not alter the CXCR7 expression in these cells (Fig. 3). Similar results were obtained in the K562, KG1a, HL-60, and MO7e cells (data not shown).
To examine the role of CXCR7 in the migration of AML cells, we knocked down CXCR4 and CXCR7 in the U937 and MO7e cells using siRNA technology. CXCL12 siRNA significantly downregulated CXCL12 mRNA expression (up to 90%) and reduced the cell surface expression of this chemokine, as evidenced by flow cytometric analysis. The transfection of AML cells with CXCR4 siRNA, but not CXCR7 siRNA, significantly impaired the CXCL12-induced transmigration of the AML cells, indicating that CXCR7 is not involved in the CXCL12-mediated chemotaxis of these cells (Fig. 4).
To examine the roles of CXCR4 and CXCR7 in the survival and proliferation of AML cells, we knocked down CXCR4 and CXCR7 in the U937 and MO7e cells using siRNA technology. First, we examined whether CXCR7 knockdown in these cells influenced serum-deprivation-induced apoptosis. Cells were incubated in RPMI-1640 medium without FBS for up to 48 hr, stained with Annexin V, and analyzed by flow cytometry. CXCR7 knockdown did not alter the induction of apoptosis in these leukemia cells. Next, we examined whether CXCR7 knockdown in these cells influenced spontaneous cell proliferation. After the transfection of siRNAs, the cells were incubated in serum-free X-VIVO medium for up to 72 hr and subjected to the CCK-8 assay. The transfection of the AML cells with CXCR7 siRNA did not affect the proliferation of these cells (Fig. 4).
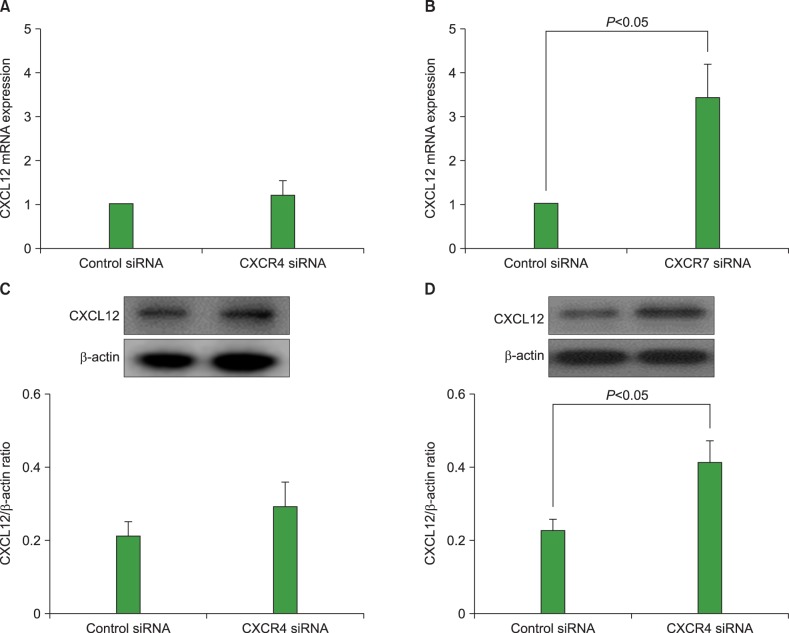
We examined whether knockdown of CXCR4 or CXCR7 in AML cells affected the expression of CXCL12 in these cells. Transfection of U937 cells with 25 nM CXCR7 siRNA upregulated the CXCL12 mRNA expression and significantly increased CXCL12 production. CXCR4 knockdown did not affect the CXCL12 expression in these cells (Fig. 5). Similar results were observed in other AML cell lines (data not shown).
All AML cell lines and primary AML cells examined in this study expressed CXCR7, both in the cytoplasm and on the cell surface at various levels, indicating that most AML cells express CXCR7. Hypoxia and ischemia are known to upregulate CXCR4 expression in most tissues as a tissue repairing mechanism [18]. In contrast, the effects of ischemia on CXCR7 expression do not seem to be uniform. For example, hypoxic injury led to enhanced CXCR7 expression in neuronal cells [19] and mesenchymal stem cells [20]; however, it did not alter the CXCR7 expression in colon cancer cells [21]. In the present study, hypoxia did not affect the CXCR7 expression in AML cells, confirming that the effects of hypoxia on CXCR7 expression differ among cell types. Various cytokines, including hematopoietic growth factors and proinflammatory cytokines, did not alter the CXCR7 expression in the AML cells. These results suggest that the regulatory mechanisms for CXCR7 are unique and different from those of CXCR4, and thus CXCR7 in AML cells has different roles from those of CXCR4.
In the present study, we found that CXCR7 was not involved in the CXCL12-mediated chemotaxis of AML cells, which has been shown in many other cell types. CXCR7 has previously been shown to enhance survival and growth in many cancer cell types. For example, in vitro and in vivo studies using prostate cancer cell lines suggested that upregulated CXCR7/RDC1 expression was associated with enhanced adhesive and invasive activities in addition to a survival advantage [11]. CXCL12 promoted the proliferation of CT26 colon cells and KEP1 mammary carcinoma cells, and this was blocked when CXCR7 was downregulated by intrakines or RNA interference, but not by CXCR4 inhibitors [10]. Additionally, CXCL12 secreted by human CD133-derived multipotent stromal cells promoted neural progenitor cell survival through CXCR7 [22]. In the present study, we demonstrated that knockdown of CXCR7 did not affect the serum-deprivation-induced apoptosis of AML cells. In addition, CXCR7 did not influence the spontaneous growth of AML cells in vitro. A recent study demonstrated a role of CXCR7, complementary to that played by CXCR4, in the control of HSC and HPC cycling, survival, and colony formation induced by CXCL12. The study also provided evidence for the involvement of β-arrestins as signaling hubs downstream of both CXCL12 receptors in primary human HPCs [23]. The implications of the differential roles of CXCR7 in cell survival and growth in normal HPCs and AML cells remain to be elucidated.
We have previously shown that AML cells produced CXCL12 and that knockdown of CXCR4 in AML cells diminished spontaneous growth of these cells in vitro, indicating that endogenous CXCL12 expression by AML cells supports their own autonomous growth via CXCR4 [24]. In the present study, knockdown of CXCR7 in AML cells did not alter the spontaneous proliferation of these cells, indicating that CXCR7 is not involved in the direct modulation of spontaneous cell growth. These results provide another example of the different functional roles of CXCR4 and CXCR7 in a specific cell-type.
Regarding the interaction between CXCR4 and CXCR7, several modes of CXCL12 signaling have been proposed. In certain cells, all effects initiated by CXCL12 are dedicated to signaling via 1 receptor; i.e., via CXCR4 or CXCR7. CXCR7, if expressed, can act as a ligand scavenger in cells that exclusively signal via CXCR4 [25]. Exclusive CXCR7 signaling can be mediated by the activation of β arrestins or, in certain cells, trimeric G proteins [26]. The possible function of co-expressed CXCR4 in such cells (which, in other cells, is signal transduction mainly via G protein activation) remains unclear. In several cell types, both receptors are active. Occasionally, CXCR4 and CXCR7 cooperate or mutually assist each other or modulate each other's functions, e.g., by heterodimerization, to mediate the effects of CXCL12 [27]. Alternatively, CXCR4 and CXCR7 can mediate different cellular responses in the same cell or can redundantly mediate identical effects via different signaling cascades [282930]. Interestingly, we observed that knockdown of CXCR7 upregulated CXCL12 expression in AML cells. These results suggest that CXCR7 is somehow involved in CXCL12 regulation and thus may affect the CXCL12/CXCR4 axis in AML cells.
In summary, we have shown that most AML cells express not only CXCR4 but also CXCR7 and that CXCR7 plays a role in the regulation of autocrine CXCL12 in AML cells.
References
1. Peled A, Petit I, Kollet O, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999; 283:845–848. PMID: 9933168.

2. Möhle R, Bautz F, Rafii S, Moore MA, Brugger W, Kanz L. The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood. 1998; 91:4523–4530. PMID: 9616148.
3. Voermans C, van Heese WP, de Jong I, Gerritsen WR, van Der Schoot CE. Migratory behavior of leukemic cells from acute myeloid leukemia patients. Leukemia. 2002; 16:650–657. PMID: 11960346.

4. Tavor S, Petit I, Porozov S, et al. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 2004; 64:2817–2824. PMID: 15087398.

5. Majka M, Rozmyslowicz T, Honczarenko M, et al. Biological significance of the expression of HIV-related chemokine coreceptors (CCR5 and CXCR4) and their ligands by human hematopoietic cell lines. Leukemia. 2000; 14:1821–1832. PMID: 11021758.

6. Dommange F, Cartron G, Espanel C, et al. CXCL12 polymorphism and malignant cell dissemination/tissue infiltration in acute myeloid leukemia. FASEB J. 2006; 20:1913–1915. PMID: 16818471.
7. Balabanian K, Lagane B, Infantino S, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005; 280:35760–35766. PMID: 16107333.

8. Sierro F, Biben C, Martínez-Muñoz L, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007; 104:14759–14764. PMID: 17804806.

9. Miao Z, Luker KE, Summers BC, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumorassociated vasculature. Proc Natl Acad Sci U S A. 2007; 104:15735–15740. PMID: 17898181.

10. Meijer J, Ogink J, Roos E. Effect of the chemokine receptor CXCR7 on proliferation of carcinoma cells in vitro and in vivo. Br J Cancer. 2008; 99:1493–1501. PMID: 18854833.

11. Wang J, Shiozawa Y, Wang J, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008; 283:4283–4294. PMID: 18057003.

12. Maussang D, Mujić-Delić A, Descamps FJ, et al. Llama-derived single variable domains (nanobodies) directed against chemokine receptor CXCR7 reduce head and neck cancer cell growth in vivo. J Biol Chem. 2013; 288:29562–29572. PMID: 23979133.

13. Iwakiri S, Mino N, Takahashi T, et al. Higher expression of chemokine receptor CXCR7 is linked to early and metastatic recurrence in pathological stage I nonsmall cell lung cancer. Cancer. 2009; 115:2580–2593. PMID: 19309748.
14. D'Alterio C, Consales C, Polimeno M, et al. Concomitant CXCR4 and CXCR7 expression predicts poor prognosis in renal cancer. Curr Cancer Drug Targets. 2010; 10:772–781. PMID: 20578990.
15. Zabel BA, Miao Z, Lai NL, et al. CXCR7 protein expression correlates with elevated mmp-3 secretion in breast cancer cells. Oncol Lett. 2010; 1:845–847. PMID: 22966392.

16. Berahovich RD, Zabel BA, Penfold ME, et al. CXCR7 protein is not expressed on human or mouse leukocytes. J Immunol. 2010; 185:5130–5139. PMID: 20889540.

17. Tarnowski M, Liu R, Wysoczynski M, Ratajczak J, Kucia M, Ratajczak MZ. CXCR7: a new SDF-1-binding receptor in contrast to normal CD34(+) progenitors is functional and is expressed at higher level in human malignant hematopoietic cells. Eur J Haematol. 2010; 85:472–483. PMID: 20887389.

18. Schioppa T, Uranchimeg B, Saccani A, et al. Regulation of the chemokine receptor CXCR4 by hypoxia. J Exp Med. 2003; 198:1391–1402. PMID: 14597738.

19. Schönemeier B, Schulz S, Hoellt V, Stumm R. Enhanced expression of the CXCl12/SDF-1 chemokine receptor CXCR7 after cerebral ischemia in the rat brain. J Neuroimmunol. 2008; 198:39–45. PMID: 18513805.

20. Liu H, Xue W, Ge G, et al. Hypoxic preconditioning advances CXCR4 and CXCR7 expression by activating HIF-1α in MSCs. Biochem Biophys Res Commun. 2010; 401:509–515. PMID: 20869949.

21. Romain B, Hachet-Haas M, Rohr S, et al. Hypoxia differentially regulated CXCR4 and CXCR7 signaling in colon cancer. Mol Cancer. 2014; 13:58. PMID: 24629239.

22. Bakondi B, Shimada IS, Peterson BM, Spees JL. SDF-1α secreted by human CD133-derived multipotent stromal cells promotes neural progenitor cell survival through CXCR7. Stem Cells Dev. 2011; 20:1021–1029. PMID: 20854168.

23. Torossian F, Anginot A, Chabanon A, et al. CXCR7 participates in CXCL12-induced CD34+ cell cycling through β-arrestindependent Akt activation. Blood. 2014; 123:191–202. PMID: 24277075.

24. de Lourdes Perim A, Guembarovski RL, Oda JM, et al. CXCL12 and TP53 genetic polymorphisms as markers of susceptibility in a Brazilian children population with acute lymphoblastic leukemia (ALL). Mol Biol Rep. 2013; 40:4591–4596. PMID: 23653000.

25. Naumann U, Cameroni E, Pruenster M, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010; 5:e9175. PMID: 20161793.

26. Rajagopal S, Kim J, Ahn S, et al. Beta-arrestin- but not G proteinmediated signaling by the "decoy" receptor CXCR7. Proc Natl Acad Sci U S A. 2010; 107:628–632. PMID: 20018651.
27. Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009; 113:6085–6093. PMID: 19380869.

28. Mazzinghi B, Ronconi E, Lazzeri E, et al. Essential but differential role for CXCR4 and CXCR7 in the therapeutic homing of human renal progenitor cells. J Exp Med. 2008; 205:479–490. PMID: 18268039.
29. Sasado T, Yasuoka A, Abe K, et al. Distinct contributions of CXCR4b and CXCR7/RDC1 receptor systems in regulation of PGC migration revealed by medaka mutants kazura and yanagi. Dev Biol. 2008; 320:328–339. PMID: 18602095.

30. Wang Y, Li G, Stanco A, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011; 69:61–76. PMID: 21220099.

Fig. 1
Acute myeloid leukemia (AML) cells express and produce C-X-C chemokine receptor 7 (CXCR7). (A) Semi-quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis in AML cell lines. (B) Western blot analysis in AML cell lines. (C) Flow cytometric analysis for CXCR7 in AML cell lines. (D) Flow cytometric analysis after dual staining of CXCR4 and CXCR7 in AML cell lines. (E) Flow cytometric analysis for CXCR7 in CD34+ cells obtained from the bone marrow of 5 patients with AML. (F) Immunofluorescence staining after permeabilization for CXCR4 and CXCR7 in U937 cells.
Abbreviations: CXCL12, C-X-C motif ligand 12; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.

Fig. 2
C-X-C motif ligand 12 (CXCL12) induces internalization of cell surface C-X-C chemokine receptor 7 (CXCR7) in acute myeloid leukemia (AML) cells. Flow cytometric analysis for cell surface CXCR7 in U937 cells before and after a 2-hr incubation with CXCL12 (200 ng/mL).

Fig. 3
Cytokines and hypoxia do not alter C-X-C chemokine receptor 7 (CXCR7) expression in acute myeloid leukemia (AML) cells. (A) AML cell lines were treated with cytokines at various concentrations for up to 72 hr and then subjected to flow cytometric analysis. Representative results (upper panel) and fold-increase of mean fluorescence intensity (MFI) of the experimental cells from that in the controls (no cytokine treatment) (lower panel) from 3 independent experiments, in which U937 cells were treated with 10 µg of each cytokine for 24 hr, are shown. Cells were incubated with 5% CO2 and 1% O2, balanced with N2 gas, at 37℃ for up to 12 hr, and then subjected to flow cytometric analysis (B) and western blot analysis (C).
Abbreviations: IL-6, interleukin 6; GM-SCF, granulocyte-macrophage colony-stimulating factor; TGF-β, tumor growth factor-β; INF-γ, interferon-γ; TNF-α, tumor necrosis factor-α; G-CSF, granulocyte colony-stimulating factor; SCF, stem cell factor; TPO, thrombopoietin; Con, control.

Fig. 4
C-X-C chemokine receptor 7 (CXCR7) does not affect migration, serum-deprivation-induced apoptosis, or spontaneous proliferation of acute myeloid leukemia (AML) cells. Using small interfering RNA (siRNA) technology, CXCR4 and CXCR7 were knocked down in MO7e and U937 cells and subsequent biological alterations occurring in the cells were evaluated. (A, B) Transmembrane migration of the cells. Data are expressed as mean±SD of the percentages of migrated cells. (C, D) Data are expressed as mean±SD of the Annexin V-positive apoptotic cells analyzed using flow cytometry. (E, F) Data are expressed as mean±SD of the relative proliferation index at indicated time. All data are from 3 independent experiments.
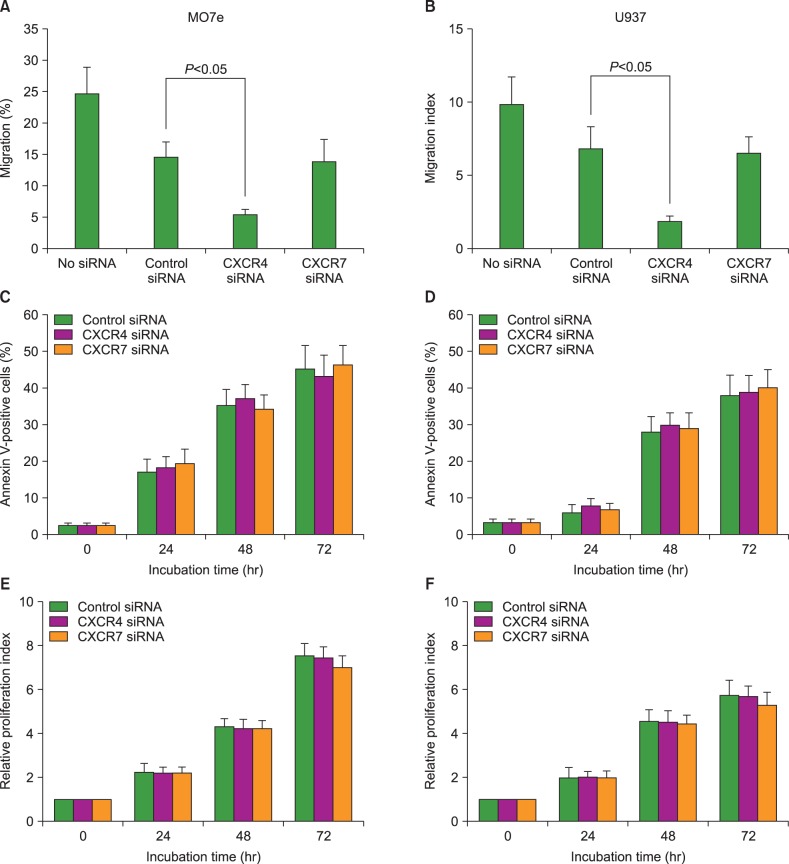
Fig. 5
Knock-down of C-X-C chemokine receptor 7 (CXCR7) upregulates C-X-C motif ligand 12 (CXCL12) expression in acute myeloid leukemia (AML) cells. Using small interfering RNA (siRNA) technology, CXCR4 and CXCR7 were knocked down in U937 cells and then subjected to quantitative reverse transcription polymerase chain reaction (RT-PCR) (A, B) and western blot analysis (C, D). Data are expressed as mean±SD of 3 independent experiments.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download