1. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998; 282:1145–1147. PMID:
9804556.

2. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. PMID:
16904174.

3. Tachibana M, Amato P, Sparman M, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013; 153:1228–1238. PMID:
23683578.

4. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. PMID:
18035408.

5. Obokata H, Wakayama T, Sasai Y, et al. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014; 505:641–647. PMID:
24476887.
6. Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012; 10:678–684. PMID:
22704507.

7. Liao SM. Rescuing human embryonic stem cell research: the blastocyst transfer method. Am J Bioeth. 2005; 5:8–16. PMID:
16282102.

8. Kim S, Izpisua Belmonte JC. Pluripotency of male germline stem cells. Mol Cells. 2011; 32:113–121. PMID:
21448589.

9. Zhou T, Benda C, Dunzinger S, et al. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012; 7:2080–2089. PMID:
23138349.

10. Vitaloni M, Pulecio J, Bilic J, Kuebler B, Laricchia-Robbio L, Izpisua Belmonte JC. MicroRNAs contribute to induced pluripotent stem cell somatic donor memory. J Biol Chem. 2014; 289:2084–2098. PMID:
24311783.

11. Seki T, Yuasa S, Oda M, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010; 7:11–14. PMID:
20621043.

12. Choi SM, Liu H, Chaudhari P, et al. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011; 118:1801–1805. PMID:
21628406.

13. Huang K, Shen Y, Xue Z, et al. A panel of CpG methylation sites distinguishes human embryonic stem cells and induced pluripotent stem cells. Stem Cell Reports. 2013; 2:36–43. PMID:
24511466.

14. Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009; 4:e7076. PMID:
19763270.

15. Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007; 448:196–199. PMID:
17597760.

16. Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010; 107:9222–9227. PMID:
20442331.

17. West FD, Terlouw SL, Kwon DJ, et al. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010; 19:1211–1220. PMID:
20380514.

18. Hamanaka S, Yamaguchi T, Kobayashi T, et al. Generation of germline-competent rat induced pluripotent stem cells. PLoS One. 2011; 6:e22008. PMID:
21789202.

19. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007; 448:313–317. PMID:
17554338.

20. Morizane A, Doi D, Kikuchi T, et al. Direct comparison of autologous and allogeneic transplantation of iPSC-derived neural cells in the brain of a nonhuman primate. Stem Cell Reports. 2013; 1:283–292. PMID:
24319664.

21. Guha P, Morgan JW, Mostoslavsky G, Rodrigues NP, Boyd AS. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013; 12:407–412. PMID:
23352605.

22. Emborg ME, Liu Y, Xi J, et al. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013; 3:646–650. PMID:
23499447.

23. Nazor KL, Altun G, Lynch C, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012; 10:620–634. PMID:
22560082.

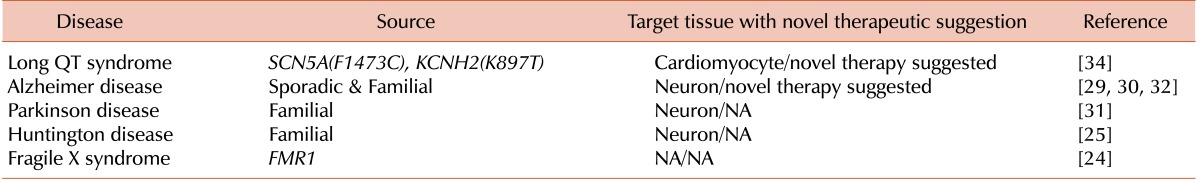
24. Eiges R, Urbach A, Malcov M, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007; 1:568–577. PMID:
18371394.

25. Bradley CK, Scott HA, Chami O, et al. Derivation of Huntington's disease-affected human embryonic stem cell lines. Stem Cells Dev. 2011; 20:495–502. PMID:
20649476.

26. Kim C, Wong J, Wen J, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013; 494:105–110. PMID:
23354045.

27. Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008; 134:877–886. PMID:
18691744.

28. Wang H, Doering LC. Induced pluripotent stem cells to model and treat neurogenetic disorders. Neural Plast. 2012; 2012:346053. PMID:
22888453.

29. Yagi T, Ito D, Okada Y, et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet. 2011; 20:4530–4539. PMID:
21900357.

30. Israel MA, Yuan SH, Bardy C, et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012; 482:216–220. PMID:
22278060.

31. Reinhardt P, Schmid B, Burbulla LF, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013; 12:354–367. PMID:
23472874.

32. Kondo T, Asai M, Tsukita K, et al. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013; 12:487–496. PMID:
23434393.

33. Patel P, Mital S. Stem cells in pediatric cardiology. Eur J Pediatr. 2013; 172:1287–1292. PMID:
23292032.

34. Terrenoire C, Wang K, Tung KW, et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol. 2013; 141:61–72. PMID:
23277474.

35. Moreno JD, Clancy CE. Pathophysiology of the cardiac late Na current and its potential as a drug target. J Mol Cell Cardiol. 2012; 52:608–619. PMID:
22198344.

36. Hwang HS, Hasdemir C, Laver D, et al. Inhibition of cardiac Ca2+ release channels (RyR2) determines efficacy of class I antiarrhythmic drugs in catecholaminergic polymorphic ventricular tachycardia. Circ Arrhythm Electrophysiol. 2011; 4:128–135. PMID:
21270101.
37. Grossmann V, Schnittger S, Poetzinger F, et al. High incidence of RAS signalling pathway mutations in MLL-rearranged acute myeloid leukemia. Leukemia. 2013; 27:1933–1936. PMID:
23535558.

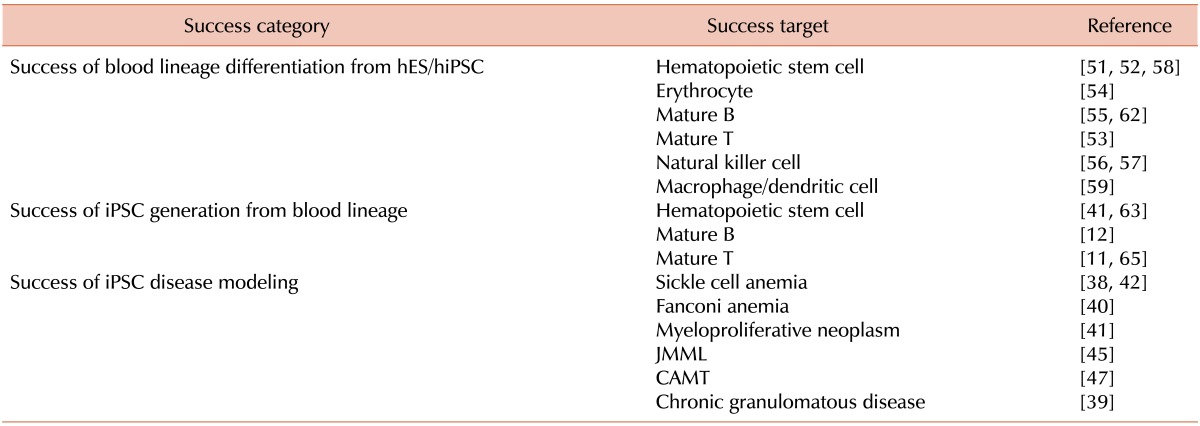
38. Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011; 118:4599–4608. PMID:
21881051.

39. Zou J, Sweeney CL, Chou BK, et al. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011; 117:5561–5572. PMID:
21411759.

40. Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009; 460:53–59. PMID:
19483674.

41. Ye Z, Zhan H, Mali P, et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009; 114:5473–5480. PMID:
19797525.

42. Sebastiano V, Maeder ML, Angstman JF, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011; 29:1717–1726. PMID:
21898685.

43. Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol Biol. 2013; 1036:81–88. PMID:
23807788.

44. Mack AA, Kroboth S, Rajesh D, Wang WB. Generation of induced pluripotent stem cells from CD34+ cells across blood drawn from multiple donors with non-integrating episomal vectors. PLoS One. 2011; 6:e27956. PMID:
22132178.

45. Gandre-Babbe S, Paluru P, Aribeana C, et al. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood. 2013; 121:4925–4929. PMID:
23620576.

46. Hirata S, Takayama N, Jono-Ohnishi R, et al. Congenital amegakaryocytic thrombocytopenia iPS cells exhibit defective MPL-mediated signaling. J Clin Invest. 2013; 123:3802–3814. PMID:
23908116.

47. Carver-Moore K, Broxmeyer HE, Luoh SM, et al. Low levels of erythroid and myeloid progenitors in thrombopoietin-and c-mpl-deficient mice. Blood. 1996; 88:803–808. PMID:
8704234.
48. Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012; 10:120–136. PMID:
22305562.

49. Kaufman DS. Toward clinical therapies using hematopoietic cells derived from human pluripotent stem cells. Blood. 2009; 114:3513–3523. PMID:
19652198.

50. Kaiser J. Gene therapy. Seeking the cause of induced leukemias in X-SCID trial. Science. 2003; 299:495. PMID:
12543948.
51. Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007; 109:2679–2687. PMID:
17148580.

52. Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2001; 98:10716–10721. PMID:
11535826.

53. Galic Z, Kitchen SG, Kacena A, et al. T lineage differentiation from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006; 103:11742–11747. PMID:
16844782.

54. Olivier EN, Qiu C, Velho M, Hirsch RE, Bouhassira EE. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp Hematol. 2006; 34:1635–1642. PMID:
17157159.

55. Wang L, Li L, Menendez P, Cerdan C, Bhatia M. Human embryonic stem cells maintained in the absence of mouse embryonic fibroblasts or conditioned media are capable of hematopoietic development. Blood. 2005; 105:4598–4603. PMID:
15718421.

56. Knorr DA, Ni Z, Hermanson D, et al. Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl Med. 2013; 2:274–283. PMID:
23515118.

57. Woll PS, Grzywacz B, Tian X, et al. Human embryonic stem cells differentiate into a homogeneous population of natural killer cells with potent in vivo antitumor activity. Blood. 2009; 113:6094–6101. PMID:
19365083.

58. Ledran MH, Krassowska A, Armstrong L, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008; 3:85–98. PMID:
18593561.

59. Zhan X, Dravid G, Ye Z, et al. Functional antigen-presenting leucocytes derived from human embryonic stem cells in vitro. Lancet. 2004; 364:163–171. PMID:
15246729.

60. Tian X, Woll PS, Morris JK, Linehan JL, Kaufman DS. Hematopoietic engraftment of human embryonic stem cell-derived cells is regulated by recipient innate immunity. Stem Cells. 2006; 24:1370–1380. PMID:
16456127.

61. Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007; 318:1920–1923. PMID:
18063756.

62. Carpenter L, Malladi R, Yang CT, et al. Human induced pluripotent stem cells are capable of B-cell lymphopoiesis. Blood. 2011; 117:4008–4011. PMID:
21343609.

63. Staerk J, Dawlaty MM, Gao Q, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010; 7:20–24. PMID:
20621045.

64. Hanna J, Markoulaki S, Schorderet P, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008; 133:250–264. PMID:
18423197.

65. Loh YH, Hartung O, Li H, et al. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010; 7:15–19. PMID:
20621044.

66. Ding Q, Lee YK, Schaefer EA, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013; 12:238–251. PMID:
23246482.

67. Gaj T, Gersbach CA, Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013; 31:397–405. PMID:
23664777.

68. Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013; 10:977–979. PMID:
23892898.

69. Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013; 14:49–55. PMID:
23169466.

70. Ramachandra CJ, Shahbazi M, Kwang TW, et al. Efficient recombinase-mediated cassette exchange at the AAVS1 locus in human embryonic stem cells using baculoviral vectors. Nucleic Acids Res. 2011; 39:e107. PMID:
21685448.






 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download