Abstract
Escherichia coli can harbor genomic pks islands that code for a polyketide-peptide genotoxin known as colibactin. E. coli strains carrying pks islands trigger genetic instability. pks islands have been significantly associated with bacteremia. We investigated the molecular epidemiology of bacteremic E. coli isolates and the prevalence of bacteremia-causing E. coli carrying pks islands. A total of 146 E. coli isolates were collected at a tertiary-care hospital from January 2015 to December 2016. The phylogenetic groups were determined by multiplex PCR. All isolates were screened by PCR for sequence type 131 (ST131)-associated single-nucleotide polymorphisms (SNPs) in mdh and gyrB. For detection of pks islands, we performed PCR for the clbB and clbN genes as colibactin system markers. Phylogenetic group B2 was the most common, accounting for 54.1% (N=79) of the isolates, followed by group D with 29.5% (N=43), group A with 11.6% (N=17), and group B1 with 4.8%. Of the group B2 isolates, 40.5% were ST131 strains and 32.9% carried pks islands. Only three ST131 isolates in group B2 carried the clbB and clbN genes, while the other 23 ST131 isolates did not. The pks gene might not be associated with ST131 strains.
Escherichia coli is one of the most common bacteremia-causing pathogens; it has several virulence factors associated with bloodstream invasion and infection [12]. Of these, the papG class II gene is thought to play a more important role in the development of E. coli bacteremia in patients with an upper urinary tract infection (UTI) than in patients with acute cholangitis [2]. However, the role of E. coli virulence factors in the pathogenesis of bloodstream infections remains unclear. Among its virulence factors, E. coli can harbor genomic pks islands that code for a polyketide-peptide genotoxin known as colibactin. E. coli strains carrying pks islands induce DNA damage and trigger genetic instability [3], and pks islands were significantly associated with bacteremia [4]. Therefore, we investigated the molecular epidemiology of bacteremic E. coli isolates and the prevalence of bacteremia-causing E. coli carrying pks islands in Korea.
A total of 146 E. coli isolates (one isolate per patient) were collected from blood samples consecutively at a tertiary-care hospital from January 2015 to December 2016. Blood cultures were incubated in the BacT/ALERT system (bioMérieux, Marcy-l'Etoile, France). Each isolate was identified using the MicroScan Walkaway (Beckman Coulter, Brea, CA, USA) or Bruker Biotyper (Bruker Daltonics, Bremen, Germany) matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry systems. Antimicrobial susceptibility testing (AST) was performed using the MicroScan Walkaway system (Beckman Coulter). The AST results were interpreted based on the CLSI guidelines [5]. Phylogenetic groups were determined by multiplex PCR using a combination of three genes (chuA, yjaA, and TSPE4.C2), as previously described; the isolates clustered into four main phylogenetic groups, A, B1, B2, and D [6]. All isolates were screened by PCR for sequence type 131 (ST131)-associated single-nucleotide polymorphisms (SNPs) in mdh and gyrB [7]. For detection of pks islands, we performed PCR for the clbB and clbN genes as colibactin system markers, as previously described [4]. The primary source of bacteremia was determined according to clinical presentation and/or evidence of an identical strain cultured near or on the same date as the onset of bacteremia. If the source of bacteremia could not be identified, it was classified as unknown origin.
Of the 146 bacteremic E. coli isolates, 107 (73.3%) were isolated from the bloodstream of patients with UTIs, and 13 (8.9%) were isolated from the bloodstream of patients with biliary tract infections (Table 1). An association with the source of infection was found for the resulting phylogenetic E. coli groups. Groups B2 and D were detected exclusively in UTIs (98/107, 91.6%), whereas group A was predominantly implicated in biliary tract infections (5/13, 38.5%). Twenty-eight isolates (group A=6, B2=14, and D=8) were recovered from patients with cancer.
Phylogenetic group B2 was the most common, accounting for 54.1% (N=79) of the isolates, followed by group D with 29.5% (N=43), group A with 11.6% (N=17), and group B1 with 4.8% (N=7; Fig. 1). Many strains belonging to group B2 are known to cause extraintestinal infections [8]. Phylogenetic analysis in a previous Korean study revealed that the majority of strains responsible for UTIs belonged to phylogenetic groups B2 (79%) and D (15%) [9]. Group B2 was also the dominant group causing bacteremia in the present study.
E. coli ST131 represents a recently emerging, globally disseminated cause of multidrug-resistant extraintestinal infections [10]. In this study, ST131 strains accounted for 24.7% (36/146) of all isolates tested. ST131 was found in groups B2 (N=32) and D (N=4), but not in groups A and B1. ST131 strains comprised 29.5% (36/122) of group B2 and D isolates.
Of the 146 bacterial isolates, 17.8% (26/146) carried pks islands, all of which belonged to group B2. Only three ST131 strains carrying pks islands were identified (2.1%). A number of studies have reported that pks islands are confined to group B2 strains [48]. Similarly, in this study, the clbB and clbN genes were detected only in group B2 isolates. This suggests that clbB is an excellent marker for group B2. Johnson et al [4] have suggested that group B2 E. coli isolates carrying pks islands increase the likelihood of bacteremia. They found that these isolates were present in 58% of blood samples, but only in 32% of fecal samples. In our study, 32.9% of the group B2 isolates from blood contained the clbB and clbN genes. Thus, the overall pks prevalence was lower than that reported in the previous study (17.8% vs 58%) [4]. We found that only three ST131 isolates in group B2 carried the clbB and clbN genes, whereas the other 23 ST131 isolates did not. These results suggest that the pks gene might not be associated with ST131 strains.
Our results describe the molecular characteristics of E. coli isolated from bloodstream infections in a Korean hospital. Approximately 54.1% of bacteremic E. coli isolates belonged to phylogenetic group B2. Of the group B2 isolates, 40.5% were ST131 strains and 32.9% carried pks islands.
Acknowledgements
This study was supported by the research fund of Hanyang University (HY-201600000002782).
References
1. Bonacorsi S, Houdouin V, Mariani-Kurkdjian P, Mahjoub-Messai F, Bingen E. Comparative prevalence of virulence factors in Escherichia coli causing urinary tract infection in male infants with and without bacteremia. J Clin Microbiol. 2006; 44:1156–1158. PMID: 16517919.
2. Wang MC, Tseng CC, Chen CY, Wu JJ, Huang JJ. The role of bacterial virulence and host factors in patients with Escherichia coli bacteremia who have acute cholangitis or upper urinary tract infection. Clin Infect Dis. 2002; 35:1161–1166. PMID: 12410475.
3. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010; 107:11537–11542. PMID: 20534522.
4. Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008; 46:3906–3911. PMID: 18945841.
5. CLSI. Performance standards for antimicrobial susceptibility testing. 25th ed. CLSI document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute;2015.
6. Doumith M, Day MJ, Hope R, Wain J, Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 2012; 50:3108–3110. PMID: 22785193.
7. Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistanturinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother. 2009; 53:2733–2739. PMID: 19398649.
8. Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006; 313:848–851. PMID: 16902142.
9. Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016; 62:84–90. PMID: 26518617.
10. Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014; 27:543–574. PMID: 24982321.
Fig. 1
Isolates carrying pks islands and those identified as strain ST131, arranged by phylogenetic group. Phylogenetic groups were determined by multiplex PCR using a combination of three genes (chuA, yjaA, and TSPE4.C2), and the isolates were clustered into four main phylogenetic groups, A, B1, B2, and D. The numbers in the dotted circle and the small dotted circle indicate ST131 positive isolates and pks-carrying isolates, respectively.
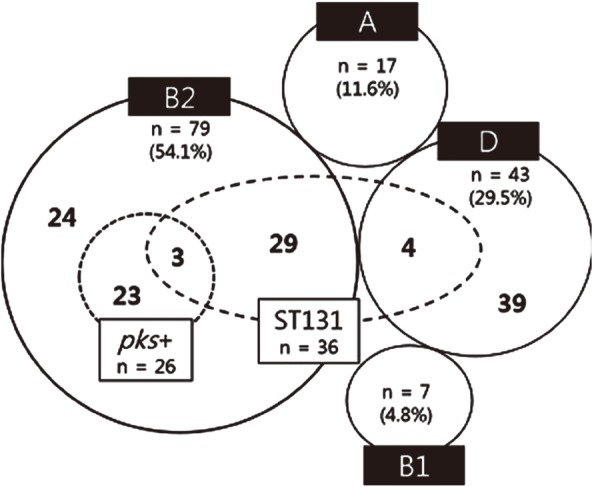
Table 1
Primary source of bacteremia classified according to phylogenetic group




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download