INTRODUCTION
Appropriate and timely antimicrobial treatment significantly improves clinical outcomes for patients with sepsis [
1]. Therefore, reducing the turn-around time of species identification (ID) and antimicrobial susceptibility test (AST) results is a high-priority topic for clinical microbiologists. The introduction of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has allowed pathogen species ID within a few minutes, as opposed to the several days required by conventional biochemical methods [
2]. Unlike other clinical samples, positive blood cultures need at least one additional day to yield pure bacterial colonies for subsequent tests and procedures. Therefore, various blood sample preparation protocols for MALDI-TOF MS have been developed to remove human cells and proteins from the blood culture broth [
34567]. However, these sample preparation methods negate the main advantages of this technique—convenience and speed—by introducing additional labor-intensive and expensive steps, and this drawback has limited their application in routine clinical microbiology practice.
To overcome the drawbacks of the current sample preparation methods, we developed an easier and cheaper method, designated the “short-term incubation method on a solid medium.” This method could eliminate the need for centrifugation and additional chemicals to extract bacterial proteins. This protocol involves a 6-hour incubation of a positive blood culture broth with subsequent ID and AST using MALDI-TOF MS and automated AST devices. This is the first prospective study to evaluate the performance of this short-term incubation method for both species ID and AST. This study also compares the performance of two rapid ID platforms using this approach: MicroFlex LT (Bruker Daltonics, Bremen, Germany) and Vitek-MS (bioMeriéux, Marcy l'Etoile, France).
METHODS
1. Blood culture samples
We used blood culture bottles inoculated with blood drawn from a peripheral vein that were referred to the laboratory in Severance hospital, Seoul, Korea, during the period from April to August 2014. Any sample with growth of more than one bacterial species or yeast was excluded from the study. Among the 334 blood cultures referred, 254 mono-microbial aerobic samples were ultimately identified and included in the study. The BacT/ALERT® 3D system (bioMeriéux, Marcy l'Etoile, France) was used for initial testing of the blood cultures. When a blood culture bottle showed a positive signal, Gram staining was performed.
Parallel to the conventional workflow shown in
Fig. 1, broth was collected from the positive blood culture bottles and inoculated on blood agar plates (BAP; Asan Pharmaceutical Co., Ltd., Seoul, Korea) and MacConkey agar plates (Becton Dickinson, Sparks, MD, USA), followed by incubation at 35℃ under 5% CO
2 atmosphere. One BAP was used for the rapid ID and AST, and the others were used for conventional biochemical ID and AST. This study was approved by the Institutional Review Board of Severance Hospital, Yonsei University Health System (2017-2648-001).
Fig. 1
Workflow of rapid and conventional species identification (ID) and antimicrobial susceptibility test (AST).
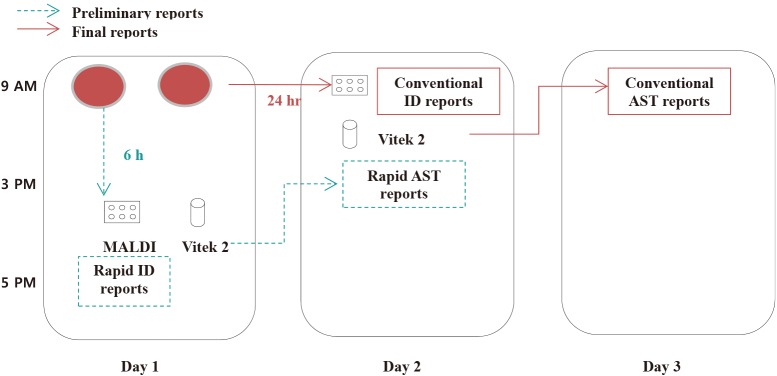

2. Conventional workflow of positive blood cultures
When the BacT/ALERT® 3D System showed a positive signal, Gram staining was performed, followed by subculture on an appropriate solid agar medium. Following overnight incubation, the colonies grown on the agar plates were used for ID and AST using the commercial automated Vitek2 system (bioMeriéux). The ID and AST results obtained using this conventional workflow were used as the standard for comparison.
3. Rapid ID and AST using the short-term incubation method
Rapid ID was performed using two MALDI-TOF MS systems: MicroFlex LT and Vitek-MS. The Vitek2 system was used for the rapid AST.
Bacterial colonies that grew on the BAP after the 6-hour incubation period were transferred to the MicroFlex LT and Vitek-MS target plates using a 1-µL sterile plastic loop. The target plates were overlaid with 1 µL of a matrix solution composed of saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/2.5% trifluoroacetic acid or with the VitekMS-CHCA matrix solution (α-cyano-4-hydroxycinnamic acid solution, ethanol, acetonitrile, solvent) for MicroFlex LT or Vitek-MS, respectively. The plates were dried in room air and subsequently subjected to MALDI-TOF MS. The ID process was performed only once for each sample. Criteria for reliable ID included confidence scores of ≥1.7 for MicroFlex LT and ≥90% for Vitek-MS [
89] according to the manufacturers' guidelines. When Vitek-MS provided more than one result with the same genus, we assumed that the result represented a reliable ID at the genus level.
To compare the AST results obtained using the rapid AST and standard methods, the minimum inhibitory concentrations obtained by both methods were categorized as susceptible, intermediate, or resistant, according to the interpretive criteria of the Vitek2 system following the CLSI recommendation [
10]. The comparison between the rapid and standard methods was classified as: agreement, very major error (false susceptibility), major error (false resistance), or minor error (susceptible/resistant vs intermediate susceptibility). AST was performed for 232 isolates, which could be possible pathogens considering the patients' clinical characteristics.
4. Time to ID
We recorded the turn-around time for both the conventional and rapid ID methods from the time of sample registration to result reporting through the five-month study duration from April to August 2014. The mean difference was calculated based on records from the laboratory information system.
5. Statistical analysis
Chi-square test was used to compare proportions. Statistical analyses were performed using a software package (Analyse-it, version 3.90.7). A P value less than 0.05 was considered statistically significant.
DISCUSSION
The aim of the present study was to evaluate the performance of a short-term incubation method for identifying bacterial pathogens from blood culture for both ID and AST, and to compare its results with those by the MicroFlex LT and Vitek-MS systems.
Bacteremia results in a high crude (24%) and attributable (17%) mortality rate [
11]; therefore, appropriate and timely antimicrobial treatment is crucial for improving the clinical outcomes of the patients with bacteremia [
1]. To eliminate the time-consuming step of conventional blood culture processing, several approaches, including MALDI-TOF MS, have been evaluated for achieving rapid pathogen ID from a positive blood culture broth [
1213]. However, the complicated sample handling steps have hampered the widespread usage of MALDI-TOF MS in routine blood culture practice. To overcome this limitation, several methods for preparation of blood culture broth have been introduced to enable the direct identification of bacteria in positive blood culture bottles, such as lysis-filtration [
3], lysis extraction [
4], protein extraction [
5], red blood cell lysis [
6], and modified lysis with nylon mesh [
7]. These methods could provide a positive ID for 91.4–99% of Gram-negative isolates and for 67.7–97% of Gram-positive isolates in about 15–45 minutes. However, all these methods require special reagents and a centrifugation process. In the short-term incubation method developed in this study, the additional preparatory steps are minimized to the preparation of one BAP for one drop of a sample.
Although this proposed rapid ID and AST method has proven to be a very powerful and simple tool, there are some drawbacks that need to be overcome [
1415]. According to Kohlmann et al [
14], rapid ID results obtained using this method allowed for optimized treatment to be recommended in 51.1% of cases, whereas only 26.4% of the Gram stain results allowed for proper treatment. Zebbe et al [
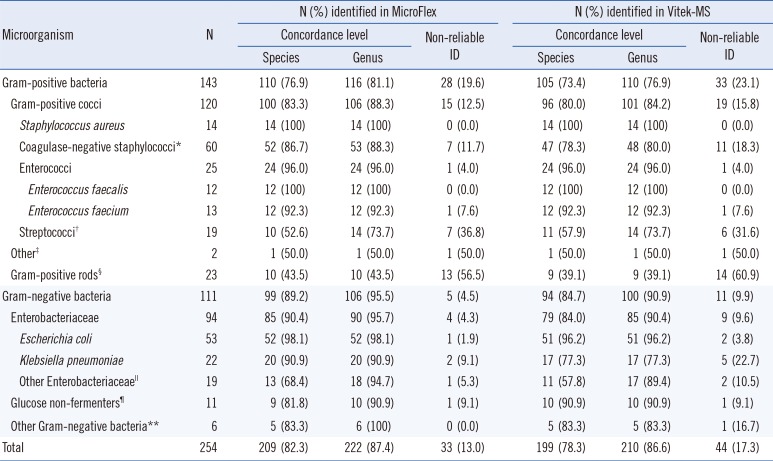
15] reported that 77% of bacteria were correctly identified with only a three-hour incubation of one drop from a positive blood culture bottle on a chocolate agar plate. Similar results were obtained in our study, in that the concordance rate was higher in Gram-negative isolates than in Grampositive isolates. The higher percentage of non-reliable IDs in Gram-positive isolates could have resulted from the relatively less growth of these organisms after the six-hour incubation.
We incubated the subculture plates for six hours because this was previously shown to substantially increase the species-level ID concordance rate of the short-term incubation method to 90.9% (159/175) from 80.6% (141/175) obtained using a fourhour incubation period [
16]. One study showed that the incubation time could be shortened by 3–6 hours using pre-warmed chocolate agar to obtain a similar correct identification rate [
17]. However, that study used a pellet streak, which requires an additional centrifugation step to obtain the pellet from the blood culture broths. We did not adopt this process because this could represent a hurdle for routine practice.
Interestingly, most of the results that were discordant between MALDI-TOF MS and the conventional ID method were due to non-reliable results rather than misidentifications. Therefore, when samples with non-reliable results were excluded, very high concordance rates for species-level ID were obtained: 94.6% (209/221) for MicroFlex LT and 94.8% (199/210) for Vitek-MS. Surprisingly, low concordance rates of 52.6% (10/19) and 57.9% (11/19) were obtained for
Streptococcus spp. with the MicroFlex LT and Vitek-MS platforms, respectively. This finding is in contrast with previous data, as summarized in a review article [
18]. The cause of this discrepancy might be the difference in incubation times or, possibly, the lack of optimal conditions for the growth of streptococci on BAPs, which could result in a failure to produce sufficient proteins for accurate MALDI-TOF MS analysis. However, given the small number of streptococci isolates in this study, the results for streptococcal species identification might be considered preliminary when specifically using the short-term incubation method on a solid medium presented herein.
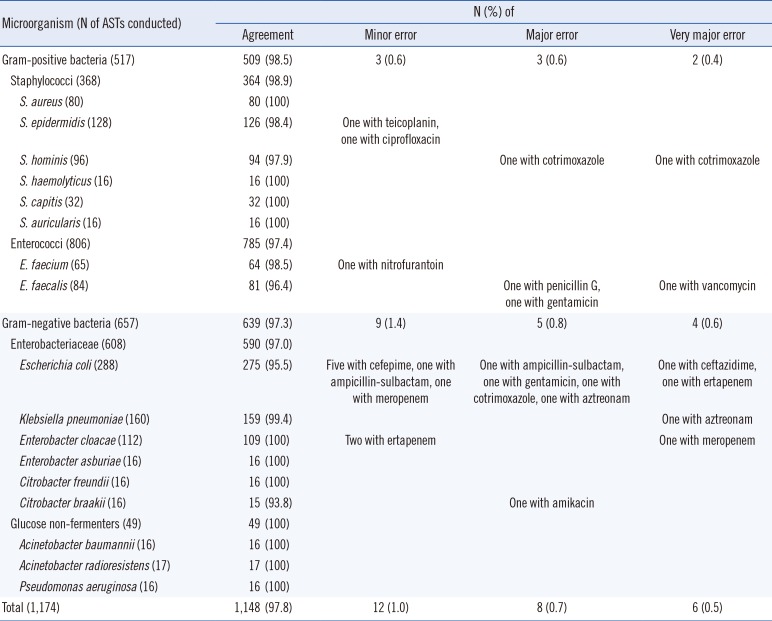
Regarding the applications of this rapid procedure for AST, the error rate was not substantial, with 1.0% minor error, 0.7% major error, and 0.5% very major error; the results were within the <3% and <1.5% US Food and Drug Administration (FDA) limits for major and very major errors, respectively [
19]. The AST results for
E. coli showed the largest number of discordances, which is likely due to the fact that the number of antimicrobials tested in
E. coli (average, 275) was much higher than that tested for the other species (average, 68). Despite this higher rate of discordance, the major and very major error rates (1.4% [4/288] and 0.7% [2/288], respectively) for
E. coli were still within the FDA limits. According to the comparison of all AST results, the concordance rates between the conventional AST methods and the short-term incubation method presented herein were very high.
One possible limitation of this study is the restricted spectrum of bacterial species analyzed, which was due to the limited sample size obtained from a clinical microbiology laboratory. Thus, these results might not apply to other laboratories. Further study using a larger number of more diverse samples over a longer period is therefore warranted to validate these results.
In conclusion, the short-term incubation on solid medium method for positive blood cultures shows acceptable performance for routine ID and AST, especially in clinically relevant bacteria. This simple and rapid method could facilitate the implementation of rapid ID and AST into the routine workflow of clinical microbiology laboratories, which could improve patient outcomes.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download