Abstract
Background
Diarrhea has been the second leading cause of death among children under the age of five, and the rapid and accurate pathogen diagnosis in patients with diarrhea is crucial for reducing morbidity and mortality. A newly developed one-step multiplex real-time PCR assay, the Allplex GI-Virus Assay, was evaluated for its ability to detect six diarrhea-causing viruses (rotavirus, norovirus genogroup I (GI) and genogroup II (GII), enteric adenovirus, astrovirus, and sapovirus) in stool samples.
Methods
The performance of the Allplex assay was compared with those of another multiplex PCR assay (Seeplex Diarrhea-V Ace Detection) and genotyping by sequencing, using 446 stool samples from patients with acute gastroenteritis.
Results
The overall agreement rates between the results of the Allplex and Seeplex assays were 98.7% for rotavirus, 99.1% for norovirus GI, 93.3% for norovirus GII, 98.0% for adenovirus, and 99.6% for astrovirus. The overall agreement rates between the Allplex assay and genotyping were 99.1% for rotavirus, 99.1% for norovirus GI, 98.7% for norovirus GII, 89.7% for adenovirus, 98.2% for astrovirus, and 99.8% for sapovirus. In addition, eight rotavirus genotypes, three norovirus GI genotypes, four norovirus GII genotypes, eight adenovirus genotypes, two astrovirus genotypes, and two sapovirus genotypes were detected.
Diarrhea is the second leading cause of death among children under the age of five, accounting for one in nine child deaths worldwide [1]. Thus, the rapid and accurate diagnosis of the underlying pathogen in patients with diarrhea is important for establishing good clinical practices aimed at reducing morbidity and mortality. Multiplex polymerase chain reaction (PCR)-based methods have been used clinically; these methods have the advantages of shorter duration of process and improved sensitivity[2345].
The Allplex GI-Virus Assay (Seegene, Seoul, Korea) is a recent one-step multiplex real-time reverse transcription PCR (qRT-PCR) assay that uses a multiple detection temperature technique (MuDT) [6] for simultaneously detecting rotavirus, norovirus genogroup I (GI) and genogroup II (GII), adenovirus type 40/41, astrovirus, and sapovirus. Compared with the previous multiplex RT-PCR assay, the Allplex assay can additionally detect sapoviruses and quantify six viral targets in a single fluorescence channel without a melting curve analysis.
This study aimed to evaluate, for the first time, the clinical performance of the Allplex assay for detecting these six diarrheacausing viruses and to compare the results with those of another multiplex PCR assay and viral genotyping.
This study was approved by the Institutional Review Board of the Hallym University Dongtan Sacred Heart Hospital (IRB number 2015-475), Korea. This study utilized stool samples collected and stored in Dongtan Sacred Heart Hospital from December 2013 to September 2016. The sample collection period was different for each virus because the positive rates for norovirus GI, adenovirus, astrovirus, and sapovirus were very low; thus, the collection period was extended to obtain an adequate number of positive samples. The number of samples and collection periods were as follows: 1) 301 leftover samples after testing for rotavirus, norovirus, adenovirus, and astrovirus from November 2015 to December 2015; 2) 56 rotavirus-positive samples from December 2015 to March 2016; 3) 43 adenovirus-positive samples from August 2015 to June 2016; and 4) 46 astrovirus-positive samples from December 2013 to March 2016. Samples with inadequate volume for subsequent testing were excluded. During the above mentioned periods, the leftover samples after testing were collected and stored at −70℃ until further analysis.
Nucleic acids were extracted from the samples, using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and a QIAcube platform (Qiagen) after the stored samples were thawed.
For the Allplex assay, 5 µL of RNA extract was mixed with 20 µL of master mix and qRT-PCR was performed using a CFX96 system (Bio-Rad, Hercules, CA, USA) under the following conditions: reverse transcription at 50℃ for 20 minutes, denaturation at 95℃ for 15 minutes, and 45 cycles of PCR (10 seconds at 95℃, 1 minute at 60℃, and 30 seconds at 72℃). All procedures were performed according to the manufacturer's instructions.
The Seeplex Diarrhea-V ACE Detection assay (Seegene) is a multiplex PCR system for detecting astrovirus, rotavirus, enteric adenovirus, and norovirus GI and GII. For this assay, following cDNA synthesis, 3 µL of cDNA was mixed with 17 µL of master mix and PCR was performed using SimpliAmp Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: 15 minutes at 94℃; followed by 40 cycles of 94℃ for 30 seconds, 60℃ for 90 seconds, and 72℃ for 90 seconds; and a final single incubation at 72℃ for 10 minutes. All procedures were performed according to the manufacturer's instructions.
Genotyping targeting the capsid genes was conducted by previously described PCR and sequencing methods [78]. Briefly, rotavirus G (VP7) and P (VP4) genotyping was performed according to the WHO manual, using specific primer sets [9], with some modifications. Norovirus capsid genotyping was performed using specific primer sets [8], and adenovirus capsid hexon genotyping was performed by PCR and sequencing using a specified primer set (ADHEX1F/AD2) or a different primer set (AD1/AD2) [10]. Additionally, astrovirus and sapovirus genotyping was performed by PCR and sequencing techniques using specific capsid primer sets [1112]. The PCR products were visualized by electrophoresis on an agarose gel and analyzed by DNA sequencing. The nucleotide sequences were analyzed using ABI Prism BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA, USA), and the genotypes were confirmed using the NCBI BLAST server of the GenBank database.
To determine the analytical sensitivity (the lower limit of detection) for norovirus GI and GII and adenovirus, commercialized virus culture fluids were purchased (Catalog #0810086CF, norovirus GI [recombinant]; #0810087CF, norovirus GII [recombinant]; and #0810085CF, adenovirus type 41; ZeptoMetrix Corporation, Buffalo, NY, USA). Viral nucleic acids were extracted using the QIAamp DSP DNA Mini Kit (Qiagen). To determine the lower limit of detection for rotavirus astrovirus, and sapovirus, in vitro transcribed RNAs were prepared using the MEGAscript T7 Kit (Life Technologies, Carlsbad, CA, USA). The prepared nucleic acids were serially diluted, and the Allplex assay was carried out using a CFX96 system (Bio-Rad). The lower limit of detection was defined as the lowest concentration that was detected in ≥95% of the replicates [13].
The analytical specificity (cross-reactivity) of the Allplex assay was assessed using 169 different pathogens (43 viruses and 126 bacteria), including 24 target viruses (see Supplemental Data Table S1). Each pathogen was tested thrice, using the same procedures for sample processing.
Inter-rater agreement statistics (Cohen's kappa) were used to compare the results of the Allplex assay, Seeplex assay, and genotyping. The kappa value was interpreted as follows: <0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, good agreement; and 0.81–1.00, very good agreement [14]. MedCalc version 15 (MedCalc Software, Mariakerke, Belgium) was used for all statistical analyses.
A total of 446 stool samples were tested, and the results of the three assays are summarized in Table 1. The overall agreement rates between the three assays for rotavirus, norovirus GI, norovirus GII, adenovirus, and astrovirus were 98.7%, 99.1%, 93.0%, 89.2%, and 97.8%, respectively. The overall agreement rates between the Allplex and Seeplex assays were 98.7% for rotavirus, 99.1% for norovirus GI, 93.3% for norovirus GII, 98.0% for adenovirus, and 99.6% for astrovirus. Furthermore, the overall agreement rates between the Allplex assay and genotyping were 99.1% for rotavirus, 99.1% for norovirus GI, 98.7% for norovirus GII, 89.7% for adenovirus, 98.2% for astrovirus, and 99.8% for sapovirus.
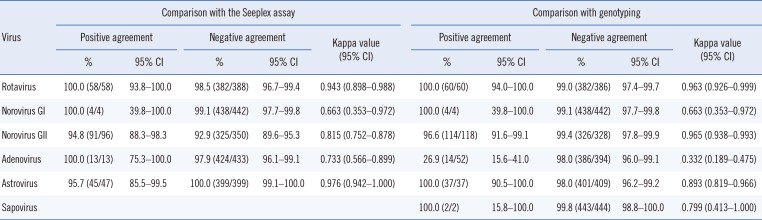
As shown in Table 2, the positive agreement rate of the Allplex assay with the Seeplex assay ranged from 94.8% to 100%, while the negative agreement rate ranged from 92.9% to 100%. The kappa correlation ranged from 0.663 to 0.976 and showed good to very good agreement. The positive agreement rate between the Allplex assay and genotyping was 100% for rotavirus, norovirus GI, astrovirus, and sapovirus; 96.6% for norovirus GII; and 26.9% for adenovirus. Moreover, the negative agreement rate ranged from 98.0% to 99.8%. The kappa correlation ranged from 0.663 to 0.965 and showed good to very good agreement, except for adenovirus (with a kappa correlation of 0.332, indicating fair agreement).Detailed results for the three methods for each virus are shown in see Supplemental Data Tables S2, S3, S4, S5, S6, S7, S8.
For rotavirus, two cases (0.4%) were negative only in the Seeplex assay, and four cases (0.9%) were positive only in the Allplex assay (see Supplemental Data Tables S2, S8). All cases had high threshold cycle (CT) values of >30 in the Allplex assay. They were interpreted as discrepancies near the cutoff with low rotavirus concentration.
For norovirus GI, four cases (0.9%) were detected by the Allplex assay only. These cases had very high CT values (>35) and were thus interpreted as discrepancies near the cutoff with very low norovirus concentration (see Supplemental Data Tables S3, S8).
For norovirus GII, 24 cases (5.4%) were negative only in the Seeplex assay, one case (0.2%) was negative only in genotyping, one case (0.2%) was positive only in the Allplex assay, four cases (0.9%) were negative only in the Allplex assay, and one case (0.2%) was positive only in the Seeplex assay (see Supplemental Data Tables S4, S8). Among the 24 cases that were negative only in the Seeplex assay, 22 had CT values >30 and were considered to have a low norovirus concentration. However, the CT values of the remaining two cases were 23.79 and 25.09, and they were identified as type GII.3. The one case that was negative only in genotyping had a CT value of 31.09 in the Allplex assay and showed weak positivity in the Seeplex assay; this case was considered to have a low norovirus concentration. The one case that was positive only in the Allplex assay had a high CT value of 35.35. The four cases that were negative only in the Allplex assay were all identified as type GII.4 by genotyping; it was estimated that the Allplex assay could not detect these cases. Moreover, the one case that was positive only in the Seeplex assay was negative by ELISA antigen test and was thus suspected to be a false positive result.
For adenovirus, two cases (0.4%) were negative only in the Seeplex assay, one case (0.2%) was negative only in genotyping, seven cases (1.6%) were positive only in the Allplex assay, and 38 cases (8.5%) were positive only in genotyping (see Supplemental Data Tables S5, S8). The two cases that were negative only in the Seeplex assay had very high CT values of 36.47 and 36.58, and the one case that was negative only in genotyping had a very high CT value of 38.97. The seven cases that were positive only in the Allplex assay had very high CT values of >35. These 10 discrepant cases were considered as low-concentration samples.
For astrovirus, eight cases (1.8%) were negative only in genotyping, and two cases (0.4%) were positive only in the Seeplex assay (see Supplemental Data Tables S6, S8). The eight cases that were negative only in genotyping consisted of genotypes that could not be detected by the genotyping method performed in our laboratory. The two cases that were positive only in the Seeplex assay showed weak bands and were interpreted as a discrepancy near the cutoff with low concentration.
For sapovirus, one discrepant case was positive in the Allplex assay, but was negative in genotyping (see Supplemental Data Tables S7, S8).
Eight rotavirus genotypes (G1P[8], G2P[4], G3P[8], G4P[6], G8P[8], G9P[4], G9P[8], and G8P[4]), three norovirus GI genotypes (GI.1, GI.5, and GI.6), four norovirus genotypes GII (GII.3, GII.4, GII.6, and GII.17), eight adenovirus genotypes (C1, C2, B3, C5, C6, A12, A31, and F41), two astrovirus genotypes (types 1 and 5), and two sapovirus genotypes (GII and GIV) were detected (see Supplemental Data Tables S2, S3, S4, S5, S6, S7, S8).
The lower limits of detection for norovirus GI (recombinant), norovirus GII (recombinant), and adenovirus using commercialized virus culture fluids were 75, 5, and 0.5 TCID50 (50% tissue culture infective dose)/mL, respectively. The lower limits of detection for rotavirus, astrovirus, and sapovirus using in vitro transcribed RNA were 5×101, 5×102, and 5×103 copies/reaction, respectively. The analytical specificity showed 100% negative signals for the 145 non-target pathogens and positive signals for the 24 target pathogens (see Supplemental Data Table S1).
Multiplex qRT-PCR assays have recently been applied for the detection of enteric viruses associated with gastroenteritis. These tests have enabled rapid, accurate, and simultaneous detection of enteric viruses with enhanced sensitivity and specificity [151617]. We evaluated the ability of a new multiplex qRT-PCR assay, the Allplex assay, to detect six common diarrhea-causing viruses and compared the results with those of the Seeplex assay and genotyping. The agreement rates among the three assays were very high. The overall agreement rates between the Allplex and Seeplex assays were ≥98.0%, except for norovirus GII (93.3%), and those between the Allplex assay and genotyping were ≥98.0%, except for adenovirus (89.7%). The low agreement rate for adenovirus was probably because the Allplex assay was designed to detect only types 40 and 41, whereas genotyping is able to detect all types of adenovirus. Therefore, 38 cases were adenovirus-positive only by genotyping, and they were all genotypes other than those of adenovirus F40 and F41.
The Allplex assay showed higher positive rates than the Seeplex assay (see Supplemental Data Tables S2, S3, S4, S5, S6, S7). The majority of the discrepant results were most likely due to low virus concentrations near the cutoff because most of them showed high CT values in the Allplex assay. Moreover, most of the discrepant results with low concentrations were negative only in the Seeplex assay, but positive in the Allplex assay and genotyping. Therefore, the most discrepant cases with low concentrations were considered as true pathogens in gastroenteritis patients.
The Seeplex assay norovirus GII-negative cases were thought to contain viral concentrations too low to be detected using the Seeplex assay, while the Allplex assay norovirus GII-negative cases were most likely caused by issues with primer and reaction conditions.
In addition, we performed genotyping by PCR and sequencing for all samples that were positive in the Allplex or Seeplex assays. To the best of our knowledge, this is the first report to evaluate the ability to detect viral genotypes by a multiplex qRT-PCR assay for the six most common diarrhea-causing viruses. Furthermore, the assays revealed common viral genotypes in the stool samples. The 38 cases not detected by the Allplex assay included genotypes 1 (5 cases), 2 (9 cases), 3 (17 cases), 5 (3 cases), 6 (2 cases), 12 (1 case), and 31 (1 case) (see Supplemental Data Table S5). While most previous studies have focused only on enteric genotypes 40 and 41 in gastroenteritis, a recent study [10] reported that other genotypes (1, 2, 3, 5, 6, 12, 31, and 55) could also be associated with enteric symptoms. That study also found that genotypes 1 and 31 were significantly associated with intussusception. Therefore, although adenovirus genotype 41 is the most common genotype isolated in the Korean patients with acute gastroenteritis, an assay for detecting other genotypes may be needed.
The Allplex assay can detect sapovirus, which cannot be detected by the Seeplex assay. Sapovirus is considered a common cause of gastroenteritis in young children less than five years of age. Of the five sapovirus genogroups, GI, GII, and GIV are thought to cause disease in humans [3]. Using the Allplex assay, we were able to detect three cases of sapovirus (see Supplemental Data Table S7) in newborn, 2-year-old, and 7-year-old patients. The one case that was positive in the Allplex assay, but negative in genotyping, had a very high CT value (36.99) and was considered as a low viral concentration sample. Therefore, the Allplex assay could detect the causative virus in pediatric gastroenteritis patients.
The Allplex assay can detect multiple targets and quantify each target in a single fluorescence channel without a melting curve analysis. Moreover, in contrast to the Seeplex assay and other previously developed assays, the Allplex assay can also detect sapoviruses.
The Allplex assay could detect six enteric viruses simultaneously in a single reaction tube and showed high agreement with the Seeplex and genotyping results. This assay enabled detection of the frequent genotypes of six enteric viruses in 2015–2016 in Korea. Therefore, the Allplex assay may constitute a good alternative method for diagnosing gastrointestinal virus infections in clinical laboratories. However, owing to genotype changes over time, continuous monitoring will be required.
Acknowledgements
We wish to thank Seegene Corporation for providing the Allplex and Seeplex assay kits and additional financial support. The funders had no role in the study design, data collection and interpretation, writing of the report, or the decision to submit the work for publication.
References
1. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012; 379:2151–2161. PMID: 22579125.
2. Liu J, Kibiki G, Maro V, Maro A, Kumburu H, Swai N, et al. Multiplex reverse transcription PCR Luminex assay for detection and quantitation of viral agents of gastroenteritis. J Clin Virol. 2011; 50:308–313. PMID: 21256076.
3. Bennett S, Gunson RN. The development of a multiplex real-time RT-PCR for the detection of adenovirus, astrovirus, rotavirus and sapovirus from stool samples. J Virol Methods. 2016; 242:30–34. PMID: 28040514.
4. Feng W, Gu X, Sui W, Zhang M, Lu B, Wang M, et al. The application and epidemiological research of xTAG GPP multiplex PCR in the diagnosis of infectious diarrhea. Zhonghua Yi Xue Za Zhi. 2015; 95:435–439. PMID: 25916780.
5. Higgins RR, Beniprashad M, Cardona M, Masney S, Low DE, Gubbay JB. Evaluation and verification of the Seeplex Diarrhea-V ACE assay for simultaneous detection of adenovirus, rotavirus, and norovirus genogroups I and II in clinical stool specimens. J Clin Microbiol. 2011; 49:3154–3162. PMID: 21775550.
6. Lee YJ, Kim D, Lee K, Chun JY. Single-channel multiplexing without melting curve analysis in real-time PCR. Sci Rep. 2014; 4:7439. PMID: 25501038.
7. Kim JS, Kim HS, Hyun J, Kim HS, Song W, Lee KM, et al. Analysis of rotavirus genotypes in Korea during 2013: an increase in the G2P[4] genotype after the introduction of rotavirus vaccines. Vaccine. 2014; 32:6396–6402. PMID: 25312273.
8. Kim JS, Kim HS, Hyun J, Kim HS, Song W. Molecular Epidemiology of Human Norovirus in Korea in 2013. Biomed Res Int. 2015; 2015:468304. PMID: 26421289.
9. WHO. Manual of rotavirus detection and characterization methods. Geneva, Switzerland: World Health Organization;2009.
10. Kim JS, Lee SK, Ko DH, Hyun J, Kim HS, Song W, et al. Associations of Adenovirus Genotypes in Korean Acute Gastroenteritis Patients with Respiratory Symptoms and Intussusception. Biomed Res Int. 2017; 2017:1602054. PMID: 28255553.
11. Zhou N, Lin X, Wang S, Wang H, Li W, Tao Z, et al. Environmental surveillance for human astrovirus in Shandong Province, China in 2013. Sci Rep. 2014; 4:7539. PMID: 25519005.
12. Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012; 50:3427–3434. PMID: 22875894.
13. Kim J, Kim H, Lee S, Oh S, Woo K, Kim S, et al. Guidelines for the Performance Evaluation of In-Vitro Diagnostic Test for the Detection of Norovirus Infection in Korea. Lab Med Online. 2017; 7:1–6.
14. Altman D. Practical statistics for medical research. London: Chapman and Hall;1991.
15. Zhang C, Niu P, Hong Y, Wang J, Zhang J, Ma X. A probe-free four-tube real-time PCR assay for simultaneous detection of twelve enteric viruses and bacteria. J Microbiol Methods. 2015; 118:93–98. PMID: 26342434.
16. Siah SP, Merif J, Kaur K, Nair J, Huntington PG, Karagiannis T, et al. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology. 2014; 46:53–59. PMID: 24300711.
17. McAuliffe GN, Anderson TP, Stevens M, Adams J, Coleman R, Mahagamasekera P, et al. Systematic application of multiplex PCR enhances the detection of bacteria, parasites, and viruses in stool samples. J Infect. 2013; 67:122–129. PMID: 23603249.
SUPPLEMENTARY MATERIALS
Supplemental Data Table S2
Comparison of the Allplex assay, Seeplex assay, and genotyping for detecting rotavirus in clinical stool specimens
Supplemental Data Table S3
Comparison of the Allplex assay, Seeplex assay, and genotyping for detecting norovirus GI in clinical stool specimens
Supplemental Data Table S4
Comparison of results for the Allplex assay, Seeplex assay, and genotyping for detecting norovirus GII in clinical stool specimens
Supplemental Data Table S5
Comparison of the Allplex assay, Seeplex assay, and genotyping for detecting adenovirus in clinical stool specimens
Supplemental Data Table S6
Comparison of the Allplex assay, Seeplex assay, and genotyping for detecting astrovirus in clinical stool specimens
Supplemental Data Table S7
Comparison of the Allplex assay and genotyping for detecting sapovirus in clinical stool specimens
Supplemental Data Table S8
Analysis of discrepant results between the Allplex assay, Seeplex assay, and genotyping
Table 1
Comparison of the Allplex assay, Seeplex assay, and genotyping for detecting rotavirus, norovirus, adenovirus, and astrovirus, and comparison of the Allplex assay and genotyping for detecting sapovirus in 446 clinical stool samples
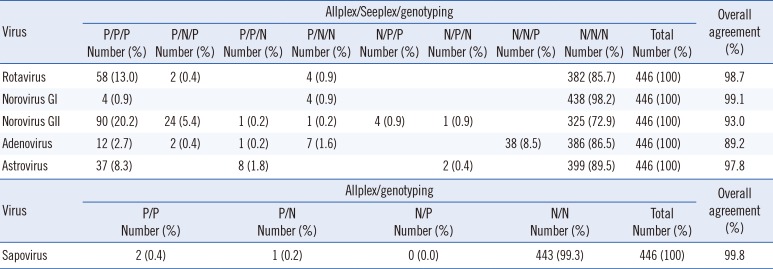
| Virus | Allplex/genotyping | Overall agreement (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P/P Number (%) | P/N Number (%) | N/P Number (%) | N/N Number (%) | Total Number (%) | ||||||
| Sapovirus | 2 (0.4) | 1 (0.2) | 0 (0.0) | 443 (99.3) | 446 (100) | 99.8 | ||||
Table 2
Comparison of the Allplex assay with the Seeplex assay and genotyping




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download