Abstract
Background
The molecular characterization of Streptococcus dysgalactiae subsp. equisimilis (SDSE) has not yet been performed in Korea. This study aimed to find the differences or similarities in the clinical features, molecular epidemiological findings, and antimicrobial resistance patterns of SDSE from two countries (Korea and Japan).
Methods
SDSE isolates were collected from Korea (N=69) from 2012–2016 and Japan (N=71) from 2014–2016. Clinical characteristics, emm genotypes, and sequence types (STs) were compared. Microdilution tests were performed using different antimicrobials, and their resistance determinants were screened.
Results
Median ages were 69 years in Korea and 76 years in Japan. The most common underlying diseases were diabetes and malignancy. Blood-derived isolates comprised 36.2% and 50.7% of Korean and Japanese isolates, respectively; mortality was not different between the two groups (5.8% vs 9.9%, P=0.53). Among Korean isolates with 20 different combined ST-emm types, ST127-stG245 (N=16), ST128-stG485 (N=10), and ST138-stG652 (N=8) were prevalent. Among Japanese isolates with 29 different combined types, ST17-stG6792 (N=11), ST29-stG485 (N=7), and ST205-stG6792 (N=6) were prevalent. Resistance rates to erythromycin, clindamycin, and minocycline were 34.8%, 17.4%, and 30.4% in Korea and 28.2%, 14.1%, and 21.4% in Japan, respectively.
Conclusions
SDSE infections commonly occurred in elderly persons with underlying diseases. There was a significant difference in the distribution of ST-emm types between the two countries. Antimicrobial resistance rates were comparable with different frequencies of resistance determinants in each country.
Streptococcus dysgalactiae subsp. equisimilis (SDSE) is a strain of large-colony forming β-hemolytic streptococci containing the Lancefield C or G antigen [12]. Rarely, this strain possesses the A antigen, which may cause misidentification if only latex agglutination is performed [34]. Although SDSE is present on the skin or mucosal surfaces, such as the respiratory tract, gastrointestinal tract, or vagina, the incidence of SDSE invasive infections has increased worldwide [2].
SDSE causes skin abscesses, cellulitis, arthritis, bacteremia, pneumonia, meningitis, or intra-abdominal abscesses; it can even cause post-streptococcal glomerulonephritis or rheumatic fever, which are the main sequelae of Streptococcus pyogenes infection [1]. Similarly, more invasive infections, such as necrotizing fasciitis or streptococcal toxic shock syndrome, may occur because of SDSE [45]. Although these two microorganisms differ with respect to their biochemical properties or presence of superantigens (i.e., streptococcal pyrogenic exotoxins), there is a large overlap of virulent factors, such as the secretion of M protein and several toxins (streptolysin S, streptolysin O, and streptokinase) into the host, or of clinical manifestations [167]. These findings indicate a close genetic relationship between these two microorganisms [289].
Therefore, clinical awareness of SDSE as a virulent pathogen is warranted. The incidence of SDSE infections seems equal to or higher than that of S. pyogenes in the United States [10] or Japan [411]. A population-based study in the United States reported that the incidence (cases per 100,000 people) of invasive β-hemolytic streptococci of groups other than A and B was 3.2 and that of S. pyogenes was 2.89 [10]. Thus, SDSE causes, in humans, a variety of diseases similar to those caused by S. pyogenes, whereas S. dysgalactiae subsp. dysgalactiae infects several different mammalian species [12].
SDSE infections are more common in elderly patients with underlying comorbidities [245711]. The emm gene, encoding the M protein, is widely used for molecular epidemiology studies of these two organisms [2]. Multilocus sequence typing (MLST) has enhanced understanding of the genetic diversity of SDSE [13]. Although SDSE is highly susceptible to β-lactam antibiotics, several reports have identified high resistance rates to classes of macrolides, lincosamides, or tetracycline [6].
A systematic review of clinical characteristics or molecular epidemiological features of SDSE has not yet been performed in Korea. We report the clinical features and molecular epidemiological findings of SDSE in Korea and compare them with those observed in Japan during similar periods. Furthermore, among erythromycin-resistant S. pyogenes isolates from two countries, the resistance determinant erm(B) was predominant in Korea, and the determinant mef(A) was frequently present in Japan [14]. Therefore, another purpose of this study was to determine the antibiotic resistance mechanisms among the SDSE isolates from the two countries.
SDSE isolates were collected using the repository at each institution from two different southern geographical areas (Gyeongsang and Jeolla provinces) in Korea between 2012 and 2016 through two tertiary-care hospitals (Gyeongsang National University Hospital [GNUH] in Gyeongsang and Chonbuk National University Hospital in Jeolla), and from three central regions (Tokyo, Saitama, and Chiba prefectures) in Japan between 2014 and 2016 through either two regional hospitals (Kitasato Medical Center [KMS] in Saitama and Mishuku Hospital supported by Federation of National Public Service and Personnel Mutual Aid Associations in Tokyo) or one laboratory center (Sanritsu Co., Ltd. in Chiba). The isolates with the possibility of true infection were consecutively collected, and repeated isolates from the same patients were excluded. The institutions in both countries sent all the isolates to a laboratory (Gyeongsang National University College of Medicine/Kitasato Institute for Life Sciences) for further genetic analysis. Bacterial identification was conducted using an API-20 Strep or Vitek-2 system (bioMerieux Inc., Marcy l'Etoile, France) and was confirmed by 16S rRNA sequencing. This identification was confirmed if the isolate sequence had ≥98.7% similarity to the 16S rRNA sequence of SDSE type strain NCFB 1356(T). All isolates were stored at −70℃ to −80℃ before being processed for further evaluation. The clinical characteristics, including underlying diseases, infection sites, surgical intervention for treatment, and mortality were reviewed using the patients' medical charts. Infection-associated death was defined as mortality within three weeks of disease onset [11].
Institutional Review Boards at the corresponding institutions (GNUH and KMS) examined and approved this study protocol (approval number GNUH 2015-10-002 and 27-58 from each institution) before starting the investigation.
The emm genotyping and MLST were described in a previous report [15]. Bacterial DNA was extracted by the boiling method (97℃ for 10 minutes) using bacterial suspension in TE buffer. Polymerase chain reaction (PCR) was performed for the emm gene with known primers, and 180 bp DNA sequences of emm genes were submitted to http://www2a.cdc.gov/ncidod/biotech/strepblast.asp to identify the genotype. Accordingly, PCR was conducted for seven housekeeping genes (gki, gtr, murI, mutS, recP, xpt, and atoB) with known primers (https://www.cdc.gov/streplab/alt-mlst-primers.html). Each sequence was amplified from purified DNA by the dye-terminator method. DNA sequences of these genes were submitted to http://pubmlst.org/sdysgalactiae/. Allele types of seven housekeeping genes were profiled to identify sequence types (STs). The STs were grouped into clonal complexes (CCs), in which the connected STs were single-locus variants differing by only one housekeeping gene [16]. Relationships between each ST in both countries were analyzed by eBURST version 3.1 (http://eburst.mlst.net/v3/). The SDSE group G strain D166B (American Type Culture Collection [ATCC] 12394) was used as a quality control.
An antimicrobial susceptibility test was performed using 14 different antimicrobial agents, including β-lactams, tetracyclines, macrolides/lincosamides, and fluoroquinolone, to evaluate antimicrobial resistance levels by broth microdilution test using either the MICroFAST or MicroScan WalkAway System (Beckman Coulter, Inc., Brea, CA, USA) according to the CLSI guidelines [1718] with the same ATCC strain (Group G strain D166B) as the quality control. In Japan, tetracycline was changed to minocycline as the screening antimicrobial in 2015. Fifty-six isolates from Japan and all Korean isolates were tested with minocycline.
Statistical analysis was performed using MedCalc Statistical Software version 17.6 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2017). The Kolmogorov-Smirnov test was used to test normal distribution. To compare two non-normally distributed groups, the Mann-Whitney U test was used. Fisher's exact probability test (two-sided) was used for categorical variables. P values<0.05 were considered statistically significant.
Demographic information and clinical manifestations of SDSE infections in Korea and Japan are shown in Table 1. We observed several significant differences between the two countries. Median age was 69 years in Korea and 76 years in Japan (P=0.0002). Surgical intervention (i.e., debridement) to treat SDSE infections was used more frequently in Korea (37.7%) than in Japan (16.9%, P=0.008). SDSE was more often isolated from pus in Korea (P=0.00005), whereas it was more frequently isolated from the sputum/throat in Japan (P=0.001). SDSE often caused infections other than sepsis, cellulitis, and septic arthritis in Japan (P=0.002). Despite these differences, mortality rates were not different between the two countries (5.8% vs 9.9%, P=0.53).
The STs and emm genotypes of SDSE isolates from Korea and Japan are shown in Tables 2 and 3, respectively. Among the 69 Korean isolates, there were 20 different combinations of ST-emm types. ST127-stG245 (N=16), ST128-stG485 (N=10), ST138-stG652 (N=8), and ST146-stG6 (N=7) were the most common types. Among the 25 blood-derived isolates, six, six, and four isolates were ST127-stG245, ST128-stG485, and ST146-stG6, respectively. A novel ST295 (allele profile, 3-3-2-2-9-22-2) was identified. The ST-emm types for four mortality cases were ST29-stC74a, ST148-stG166b, ST128-stG485, and ST138-stG652. All four strains were isolated from blood.
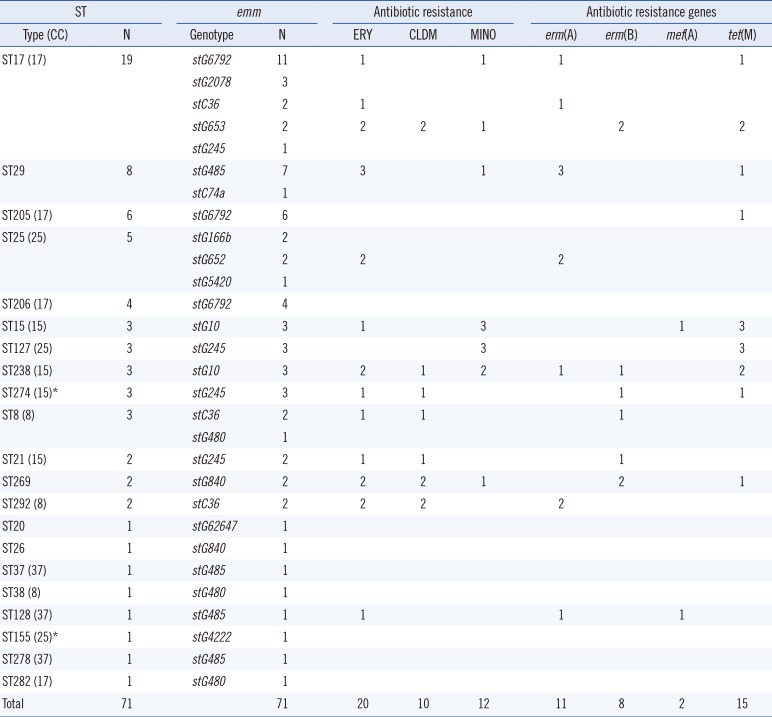
Among the 71 Japanese isolates, there were 29 different combinations of ST-emm types. ST17-stG6792 (N=11), ST29-stG485 (N=7), and ST205-stG6792 (N=6) were the most frequent types. Among the 36 blood-derived isolates, ST17-stG6792 and ST127-stG245 represented nine and three isolates, respectively. Combinations of ST-emm were more diverse for Japanese blood isolates. Seven mortality cases possessed three ST17-stG6792 and each of ST205-stG6792, ST29-stG485, ST37-stG485, and ST25-stG5420. Three strains were isolated from blood, two from open pus, one from cerebrospinal fluid, and one from sputum.
The distribution of the common emm types was quite different between the two countries. The stG245 (24.6%), stG6 (15.9%), and stG485 (14.5%) were most common in Korea, while stG6792 (29.6%), stG485 (14.1%), and stG245 (12.7%) were most frequent in Japan. The ST-relationships consisting of several CCs were observed among the Japanese isolates only. Among the Korean isolates, only CC25 was observed, whereas the Japanese isolates indicated a variety of CCs (i.e., 17, 37, 25, 15, and 8) (Figs. 1, 2).
There was no statistically significant difference in antimicrobial resistance rates between the two countries (P>0.05).
Resistance rates to erythromycin, azithromycin, clindamycin, and minocycline were 34.8%, 34.8%, 17.4%, and 30.4%, respectively, for Korean isolates (Table 2). Several ST-emm types were associated with macrolide/lincosamide and/or tetracycline resistance. Among the 24 erythromycin-resistant strains (MIC≥1 µg/mL), 12 were ST127-stG245 and six were ST128-stG485 (Table 2). Antimicrobial resistance determinants of erm(A), erm(B), and mef(A), comprised three, 14, and six, respectively. All the 12 isolates resistant to clindamycin (MIC≥1 µg/mL) contained the erm(B) gene; 10 were ST127-stG245. Among the 21 minocycline-resistant strains (MIC>4 µg/mL), seven were ST138-stG652, six were ST127-stG245, and six were ST128-stG485. Antimicrobial resistance genes of tet(M) and tet(O) comprised 20 and one, respectively. Nine strains (42.9%) resistant to tetracycline also possessed macrolide resistance determinants.
Resistance rates to erythromycin, azithromycin, clarithromycin, clindamycin, tetracycline, and minocycline were 28.2% (No. of examined isolates=71), 32.1% (N=56), 13.3% (N=15), 14.1% (N=71), 26.7% (N=15), and 21.4% (N=56), respectively, for Japanese isolates (Table 3).
Among the 20 erythromycin-resistant strains, stC36 and stG485 comprised four each (Table 3). Antimicrobial determinants of erm(A), erm(B), and mef(A) comprised 11, eight, and two isolates, respectively. There was a significant difference in the frequency of erm(A) (P=0.009) between the two countries, whereas other genes showed no significant frequency differences.
Among the 10 isolates resistant to clindamycin, erm(B) and erm(A) comprised eight and two respectively; there were three of stC36, two of stG245, two of stG840, and two of stG653. Among the 12 minocycline-resistant strains out of 56 tested, five were stG10 and three were stG245. All strains contained tet(M) except two strains with tet(O). Five strains (41.7%) resistant to tetracycline also possessed macrolide resistance determinants. One strain (ST274-stG245) isolated from blood concurrently contained erm(B), tet(M), and tet(O) genes.
The 50% minimum inhibitory concentrations (MIC50) of erythromycin, clindamycin, and minocycline were ≤0.12 µg/mL, ≤0.12 µg/mL, and 0.5 µg/mL, and the MIC90 of these antimicrobial agents were >2 µg/mL, >1 µg/mL, and >4 µg/mL, respectively, for both Korean and Japanese isolates. Fluoroquinolone-resistant isolates (N=5) were identified only in Japan. None of the tested SDSE isolates from either country were resistant to β-lactams.
Although there have been sporadic case reports regarding SDSE infections, there was no systematic report in Korea yet. There are two SDSE bacteremia cases originated from endocarditis and cellulitis, respectively, in Korea [21]. The emm types were identified as stG3251 and stG245 for these cases.
Recently, Takahashi et al [15] reported the distribution of CC17 (stG6792) SDSE harboring a complement-mediated cell lysis-like gene (sicG) in central Japan. The CC17 isolates (N=17) with sicG could be divided into three subtypes by random amplified polymorphic DNA analysis, suggesting clonal spread in central areas of Japan. In addition, there was a significant association between the presence of sicG and the development of skin/soft tissue infections.
SDSE infections seemed more invasive in Japan because the proportion of blood-derived isolates (36.2% vs 50.7%) was relatively higher in Japan. SDSE caused more wound infections in Korea (46.4% vs 11.3%) and more respiratory tract infections in Japan (4.4% vs 23.9%). The prevalence of sepsis was similar (10.2% vs 7.0%) between the two countries, whereas cellulitis was more common in Korea (44.9% vs 28.2%). More than half of the patients visited the hospitals through the emergency department. Surgical intervention was more common in Korea (37.7% vs 16.9%) because wound infections comprised a larger portion of cases. The most common underlying comorbidities in both countries were diabetes and malignancy. The higher median age (69 years vs 76 years) for Japanese patients might be the reason for the higher proportion of elderly patients in Japan. Mortality in this study was lower than that observed in other studies [11011] because we included non-invasive cases.
More diverse combinations of ST-emm types were observed in Japan (20 vs 29). Distribution of common ST-emm types was different for each country: ST127-stG245 (N=16), ST128-stG485 (N=10), ST138-stG652 (N=8), and ST146-stG6 (N=7) in Korea; ST17-stG6792 (N=11), ST29-stG485 (N=7), and ST205-stG6792 (N=6) in Japan. The following emm types also were strongly associated with bacteremia: stG245 (86%) and stG485 (60%) in Korea, and stG6792 (67%) and stG245 (67%) in Japan.
The distribution of common emm types overlapped with those from other Asian countries: stG485, stG6, and stC839 in northern Taiwan [22], and stG245, stG652, stG6, and stG485 in Beijing, China [6]. However, the distribution of virulent emm types in the United States differs greatly [10].
The prevalent invasive emm types in France (stG6, stG485, and stG6792) also overlapped with our results [23]. In contrast with results of an Austrian study, there was a relationship between emm type and infection site or invasiveness in our study [7]. Among the 11 cases with mortality, four were due to stG6792 and three were due to stG485. The stG6792 type is associated with poor prognosis (death and post-infection sequelae) in Japan [11].
Antimicrobial resistance rates significantly increased compared with those in 2007 in Korea—from 9.4% to 34.8% for erythromycin and from 3.1% to 17.4% for clindamycin [24]. Antimicrobial resistance rates to erythromycin, clindamycin, and minocycline were comparable between each country. In addition, MIC50 and MIC90 for these antibiotics were equivalent. However, the frequency of antimicrobial resistance determinants was quite different. erm(B) was most common in Korea (58.3%), whereas erm(A) was most common in Japan (55%). One isolate from Japan contained both erm(A) and mef(A). Some emm-ST or emm genotypes were associated with antimicrobial resistance patterns. stG245-ST127 was distinctively associated with resistance to both erythromycin and clindamycin in Korea. Most clindamycin-resistant strains showed higher MIC values (>2 µg/mL) for erythromycin.
Most of the minocycline-resistant isolates harbored the tet(M) gene. The tet(O) gene was very rarely present in the isolates. A recent study from central Taiwan reported a similar erythromycin resistance rate (24.0%), but a different distribution of resistance determinants was observed in this study [25]. A previous Japanese study suggested a strong association between emm types (stG245 and stG10) and erythromycin resistance [4]. However, such a phenomenon was not observed in this study. This might be because of the different sources and small number of isolates tested in this study.
In conclusion, SDSE commonly caused infections in the elderly who had diabetes or malignancy in both Korea and Japan. The proportion of bacteremia and mortality was slightly higher in Japan. The prevalent emm genotypes or STs were quite different between the two countries. Several emm or emm-ST combinations were associated with bacteremia and/or macrolide/lincosamide resistance, and this association was stronger in Korea. Resistance rates to the classes of macrolides/lincosamides and tetracycline were comparable, but the distribution of antimicrobial resistance determinants differed, indicating different resistance mechanisms. Thus, there were differences of the clinical data, epidemiological results, and antimicrobial resistance genotypes between the two countries. We should further analyze a large number of isolates from Korea and Japan and monitor these profiles sequentially.
Acknowledgments
This study was supported in part by biomedical research institute fund (GNUHBIF-2016-0003) from the GNUH, and Development Fund Foundation, Gyeongsang National University, 2015 to Sunjoo Kim. This publication made use of the Streptococcus dysgalactiae MLST website (http://pubmlst.org/sdysgalactiae/) developed at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust.
References
1. Rantala S. Streptococcus dysgalactiae subsp. equisimilis bacteremia: an emerging infection. Eur J Clin Microbiol Infect Dis. 2014; 33:1303–1310. PMID: 24682845.
2. Brandt CM, Spellerberg B. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin Infect Dis. 2009; 49:766–772. PMID: 19635028.
3. Son JA, Shin JH. Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield's group A antigen. Lab Med Online. 2013; 3:60–61. https://labmedonline.org/Synapse/Data/PDFData/9997LMO/lmo-3-60.pdf (Accessed on 20th December 2017, in Korean language).
4. Wajima T, Morozumi M, Hanada S, Sunaoshi K, Chiba N, Iwata S, et al. Molecular characterization of invasive Streptococcus dysgalactiae subsp. equisimilis, Japan. Emerg Infect Dis. 2016; 22:247–254. PMID: 26760778.
5. Bruun T, Kittang BR, de Hoog BJ, Aardal S, Flaatten HK, Langeland N, et al. Necrotizing soft tissue infections caused by Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis of groups C and G in western Norway. Clin Microbiol Infect. 2013; 19:E545–E550. PMID: 23795951.
6. Lu B, Fang Y, Huang L, Diao B, Du X, Kan B, et al. Molecular characterization and antibiotic resistance of clinical Streptococcus dysgalactiae subsp. equisimilis in Beijing, China. Infect Genet Evol. 2016; 40:119–125. PMID: 26925701.
7. Leitner E, Zollner-Schwetz I, Zarfel G, Masoud-Landgraf L, Gehrer M, Wagner-Eibel U, et al. Prevalence of emm types and antimicrobial susceptibility of Streptococcus dysgalactiae subsp. equisimilis in Austria. Int J Med Microbiol. 2015; 305:918–924. PMID: 26507866.
8. Ahmad Y, Gertz RE Jr, Li Z, Sakota V, Broyles LN, Van Beneden C, et al. Genetic relationships deduced from emm and multilocus sequence typing of invasive Streptococcus dysgalactiae subsp. equisimilis and S. canis recovered from isolates collected in the United States. J Clin Microbiol. 2009; 47:2046–2054. PMID: 19386831.
9. Jensen A, Kilian M. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol. 2012; 50:113–126. PMID: 22075580.
10. Broyles LN, Van Beneden C, Beall B, Facklam R, Shewmaker PL, Malpiedi P, et al. Population-based study of invasive disease due to beta-hemolytic streptococci of groups other than A and B. Clin Infect Dis. 2009; 48:706–712. PMID: 19187026.
11. Takahashi T, Sunaoshi K, Sunakawa K, Fujishima S, Watanabe H, Ubukata K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010; 16:1097–1103. PMID: 19732082.
12. Vandamme P, Pot B, Falsen E, Kersters K, Devriese LA. Taxonomic study of Lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996; 46:774–781. PMID: 8782689.
13. Fujita T, Horiuchi A, Ogawa M, Yoshida H, Hirose Y, Nagano N, et al. Genetic diversity in Streptococcus dysgalactiae subsp. equisimilis isolates from patients with invasive and noninvasive infections in a Japanese university hospital (2014-2015). Jpn J Infect Dis. 2017; 70:100–104. PMID: 27000456.
14. Takahashi T, Arai K, Lee DH, Koh EH, Yoshida H, Yano H, et al. Epidemiological study of erythromycin-resistant Streptococcus pyogenes from Korea and Japan by emm genotyping and multilocus sequence typing. Ann Lab Med. 2016; 36:9–14. PMID: 26522753.
15. Takahashi T, Fujita T, Shibayama A, Tsuyuki Y, Yoshida H. Prevalence of complement-mediated cell lysis-like gene (sicG) in Streptococcus dysgalactiae subsp. equisimilis isolates from Japan (2014-2016). Ann Lab Med. 2017; 37:297–304. PMID: 28445008.
16. McMillan DJ, Bessen DE, Pinho M, Ford C, Hall GS, Melo-Cristino J, et al. Population genetics of Streptococcus dysgalactiae subspecies equisimilis reveals widely dispersed clones and extensive recombination. PLoS One. 2010; 5:e11741. PMID: 20668530.
17. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 18th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute;2008.
18. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 22nd ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute;2012.
19. Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl Environ Microbiol. 2010; 76:7957–7965. PMID: 20952646.
20. Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005; 49:4798–4800. PMID: 16251336.
21. Hu JY, Kim EH, Lee HW, Cha SA, Choi JY, Kim YJ, et al. Emm typing of invasive infections caused by Streptococcus dysgalactiae. Korean J Med. 2014; 87:630–635.
22. Tseng SP, Lin YY, Tsai JC, Hsueh PR, Chen HJ, Hung WC, et al. Distribution of emm types and genetic characterization of the mgc locus in group G Streptococcus dysgalactiae subsp. equisimilis from a hospital in northern Taiwan. J Clin Microbiol. 2010; 48:2975–2977. PMID: 20573871.
23. Loubinoux J, Plainvert C, Collobert G, Touak G, Bouvet A, Poyart C, et al. Adult invasive and noninvasive infections due to Streptococcus dysgalactiae subsp. equisimilis in France from 2006 to 2010. J Clin Microbiol. 2013; 51:2724–2727. PMID: 23698531.
24. Uh Y, Hwang GY, Jang IH, Cho HM, Noh SM, Kim HY, et al. Macrolide resistance trends in beta-hemolytic streptococci in a tertiary Korean hospital. Yonsei Med J. 2007; 48:773–778. PMID: 17963333.
25. Lo HH, Nien HH, Cheng YY, Su FY. Antibiotic susceptibility pattern and erythromycin resistance mechanisms in beta-hemolytic group G Streptococcus dysgalactiae subspecies equisimilis isolates from central Taiwan. J Microbiol Immunol Infect. 2015; 48:613–617. PMID: 24856419.
Fig. 1
eBURST analysis of Streptococcus dysgalactiae subsp. equisimilis isolates (N=69) from Korea. The relationships of each ST were generated with eBURST version 3.1 (http://eburst.mlst.net/v3/).
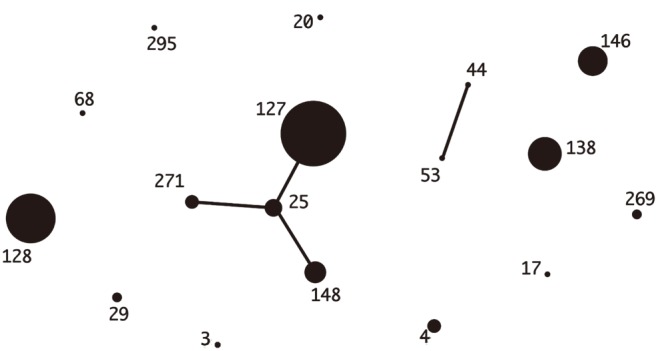
Fig. 2
eBURST analysis of Streptococcus dysgalactiae subsp. equisimilis isolates (N=71) from Japan. The relationships of each ST were generated with eBURST version 3.1 (http://eburst.mlst.net/v3/).
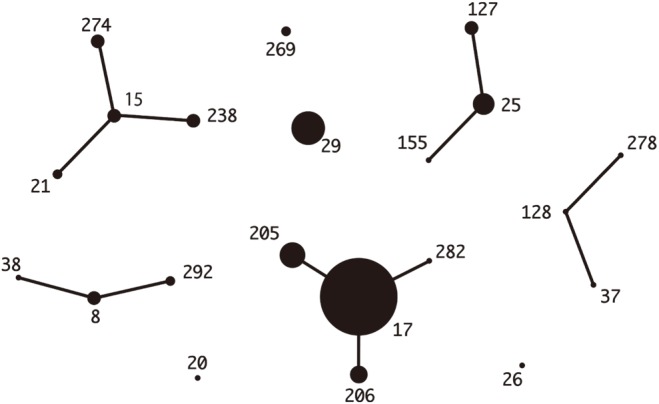
Table 1
Demographics and clinical manifestations of infectious diseases due to Streptococcus dysgalactiae subsp. equisimilis in Korea and Japan
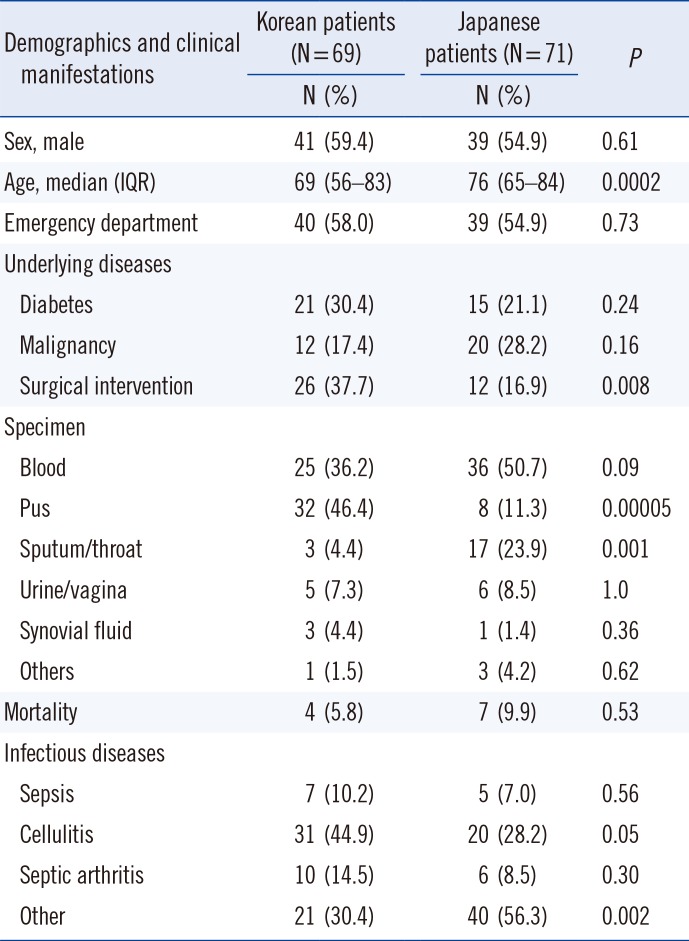
Table 2
The sequence types (STs) and emm genotypes of SDSE isolates (N=69) from Korea
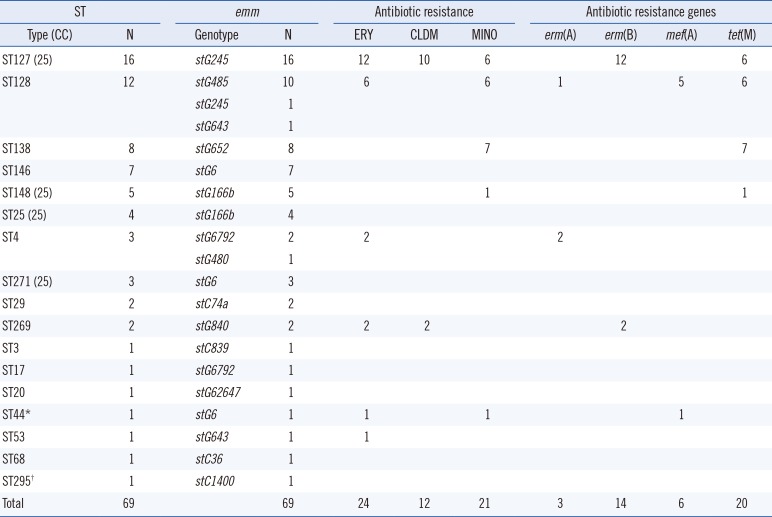
Table 3
The sequence types (STs) and emm genotypes of SDSE isolates (N=71) from Japan




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download