Abstract
In multiple myeloma (MM), hyperdiploidy (HD) is known to impart longer overall survival. However, it is unclear whether coexistent HD ameliorates the adverse effects of known high-risk cytogenetics in MM patients. To address this issue, we investigated the clinicopathological characteristics of HD with high-risk cytogenetics in MM. Ninety-seven patients with MM were included in the study. For metaphase cytogenetics (MC), unstimulated cells from bone marrow aspirates were cultured for either 24 or 48 hours. To detect HD by interphase fluorescence in situ hybridization (iFISH), we assessed trisomies of chromosomes 5, 7, 9, 11, 15, and 17. Of the 97 MM patients, 40 showed HD. The frequency of co-occurrence of HD and high-risk cytogenetics was 14% (14/97). When the clinicopathological characteristics were compared between the two groups of HD with high-risk cytogenetics vs. non-HD (NHD) with high-risk cytogenetics, the level of beta 2 microglobulin and stage distribution significantly differed (P=0.020, P=0.032, respectively). This study shows that some of the clinicopathological characteristics of MM patients with high-risk cytogenetics differ according to HD or NHD status.
Risk stratification of patients with multiple myeloma (MM) is important to predict survival and define a treatment strategy. Cytogenetic abnormalities are clinically relevant prognostic factors in MM [12]. Patients with MM can be categorized into hyperdiploidy (HD) and non-hyperdiploidy (NHD) groups according to the primary cytogenetic abnormalities. The definition of HD in MM has varied across studies; one study defined HD as numerous chromosomal trisomies and low prevalence of IgH translocations [3], and another as a chromosome count of 48–65, with a gain of at least two odd chromosomes [4]. HD in this study was defined by a chromosome count of 47 or more, with gain of one or more odd-numbered chromosomes (chromosome 3, 5, 7, 9, 11, 15, or 17). Previous studies have shown that abnormalities such as t(4;14), t(14;16), and deletion of the short arm of chromosome 17 (17p) are predictive of significantly shortened survival (defined as high-risk cytogenetics), whereas HD is associated with cytogenetics with a favorable outcome [12]. There is a small subset of patients with MM that shows evidence of both HD and high-risk cytogenetics, but its clinical significance and prognostic impact is controversial [5678].
This study aimed to investigate clinicopathological characteristics associated with HD with high-risk cytogenetics in MM, compared with those of NHD with high-risk cytogenetics in MM. Interestingly, del(5q) was detected in two patients, which corresponded to an EGR1 deletion. We examined the clinical significance of either del(5q) or an EGR1 mutation in MM and reviewed the literature.
A total of 97 patients were newly diagnosed as having MM, according to the International Myeloma Working Group diagnostic criteria [9], at two institutions in Korea (93 from Ewha Womans University Mokdong Hospital, Seoul, Korea; four from Dongguk University Ilsan Hospital, Goyang, Korea) between 2010 and 2015. The demographics, laboratory, and clinical data of the patients were retrospectively reviewed. This study was exempted from approval from the institutional review board, as it was a retrospective review of existing medical records, and all information was de-identified. Demographic and laboratory characteristics of the patients are summarized in Supplemental Data Tables S1 and S2.
HD was confirmed by metaphase cytogenetics (MC) and/or interphase fluorescence in situ hybridization (iFISH). For MC, unstimulated cells from bone marrow aspirates were cultured for either 24 or 48 hours. All data were retrospectively reviewed and reanalyzed according to the International System for Human Cytogenetic Nomenclature (ISCN 2013) guidelines [10]. iFISH was performed using the following probes (Abbott/Vysis, Downers Grove, IL, USA; Metasystems, Heidelberg, Germany; or Kreatech, Amsterdam, the Netherlands): IgH dual color break apart rearrangement probe, TP53 dual color probe, CEP7/D7S486 dual color probe, 13q14/13q34 dual color probe, IgH/CCND1 dual color probe, dual fusion probe, IgH/FGFR3 dual color probe, dual fusion probe, IgH/MAF dual color probe, dual fusion probe, 1q21/1p32 dual color probe, EGR1/D5S721,D5S23 dual color probe, p16/CEP9 dual color probe, and/or CEP15 single color probe. To detect HD by iFISH, we assessed trisomies of chromosomes 5, 7, 9, 11, 15, and 17, using targeted probes. At least 200 interphase cells from each case were counted.
Of the 97 patients with MM, 57 (59%) were classified into the NHD group, and HD was detected in 40 patients (41%) by MC and/or iFISH. At diagnosis, iFISH in two patients (See Supplemental Data Table S1; cases 6 and 9) revealed HD, whereas a normal karyotype was revealed by MC. However, after the disease progressed, HD was also detected by MC.
We defined high-risk cytogenetics as the presence of t(4;14), t(14;16), or del(17p), based on the revised International Staging System proposed by the International Myeloma Working Group [1]. Among patients newly diagnosed with MM, the frequency of co-occurrence of HD and high-risk cytogenetics was 14% (14/97); t(4;14), (14;16), and del(17p) were observed in 7% (7/97), 2% (2/97), and 8% (8/97) of the patients, respectively (Table 1). Three patients presented concurrent, multiple high-risk cytogenetics (See Supplemental Data Table S1; cases 12, 18, and 35). We also reviewed the frequencies of high-risk cytogenetics in the NHD group (See Supplemental Data Table S2) and compared them with those in the HD group. The difference in the frequencies of high-risk cytogenetics was not statistically gnificant (P=0.467) between the two groups.
Table 1 summarizes the comparative analysis between the two groups of HD with high-risk cytogenetics vs. NHD with high-risk cytogenetics.
The difference in stage distribution, determined according to the International Staging System (ISS), did not reach statistical significance (P=0.051), but stage distribution determined according to the revised International Staging System (ISS-R) [11] was significantly different between the two groups (P=0.032) (Table 1). It is noteworthy that 57% of patients with HD and high-risk cytogenetics belonged to ISS-R stage II, whereas 81% of patients with NHD and high-risk cytogenetics belonged to stage III.
Gain of chromosome 9 (22 patients) was the most commonly observed aneuploidy in the HD group, identified by iFISH, followed by gain of chromosome 5 (n=21), 15 (n=19), similar to the frequencies reported in previous studies [1213]. iFISH using probes targeting chromosomes 5, 9, and 15 for the detection of HD showed various patterns. In one patient (See Supplemental Data Table S1; case 20), iFISH showed trisomy of chromosome 5 (43.5%), whereas both trisomy and tetrasomy were observed for chromosomes 9 (trisomy, 13.0%; tetrasomy, 48.5%) and 15 (trisomy, 18.5%; tetrasomy, 22.5%). In another patient (See Supplemental Data Table S1; case 32), trisomies of chromosomes 9 (72.0%) and 11 (61.0%) were revealed by iFISH, whereas both trisomy and tetrasomy were observed for chromosome 15 (trisomy, 30.0%; tetrasomy, 29.5%). In one patient (See Supplemental Data Table S1; case 6), both trisomies and tetrasomies of chromosome 5 (trisomy 26.5%, tetrasomy 78.5%) and 9 (trisomy 14.5%, tetrasomy 73.5%) were observed at relapse by iFISH, whereas only trisomies of chromosome 5 (28.0%), 9 (77.5%), and 15 (61.5%) were observed at diagnosis.
As for chromosome 5 abnormalities, the del(5q) was observed in two patients by MC (See Supplemental Data Table S1; cases 9 and 38). One patient (case 9) showed a normal karyotype at diagnosis, but partial trisomy 5 (two normal chromosome 5s and one with a 5q deletion) was detected by MC at nine months (Fig. 1A). In another patient (case 38), del(5q) was identified by MC and 5q signal loss, corresponding to an EGR1 deletion of 5q31 (21.6%), was confirmed by iFISH (Fig. 1B and C). Furthermore, in one patient (case 40), iFISH indicated two clones: one with trisomy 5 (15.0%), and the other with partial trisomy 5 [two normal and one with del(5q)] (11.5%), whereas only trisomy 5 was revealed by MC. Two patients (cases 6 and 16) showed partial tetrasomy 5 by iFISH: three normal and one with del(5q) (78.5% in case 6; 22.0% in case 16) (Fig. 1D and E).
Similar to our findings, one study showed partial trisomy 5 (two normal and one with a 5q deletion) in clonal plasma cells of patients newly diagnosed as having MM [14]. EGR1 as a candidate gene for del(5q) in MDS has been associated with an individual's response to lenalidomide [15]. However, another study showed that an EGR1 mutation is highly associated with HD in MM [16]. In MM, EGR1 was recently shown to be involved in recruiting MYC to the promoters of NOXA and BIM and inducing p53-independent apoptosis [1718]. The clinical significance of del(5q) or an EGR1 mutation on 5q in MM remains to be ascertained.
There is no consensus on the prognostic impact or risk associated with concurrent HD and high-risk cytogenetics. A few studies have shown that the poor outcomes observed in patients with high-risk cytogenetics are not ameliorated by the co-occurrence of HD [78]. In contrast, one study showed that patients with high-risk cytogenetics have higher survival rates when trisomies are present than when they are absent [6]. Another study showed that trisomy 15 increases the rate of progression-free survival in patients with del(17p), whereas HD does not improve clinical outcomes in patients harboring t(4;14) [5]. These discrepant findings may be attributed to different treatment regimens. Certain therapies (i.e., bortezomib-based therapies) can mitigate the risk associated with specific abnormalities, as in the case of t(4;14) or del(17p) [12].
Our study showed that a considerable subset—14% (14/97)—of all MM patients had HD and coexistent high-risk cytogenetics. Of the 40 HD patients, 35% (14/40) presented high-risk cytogenetics, whereas 28% (16/57) of NHD patients presented high-risk cytogenetics. Moreover, the distribution of stages according to ISS-R differed between the groups with HD with high-risk cytogenetics and NHD with high-risk cytogenetics. This study indicates that the clinicopathological characteristics of MM patients with high-risk cytogenetics may differ according to HD or NHD status.
Further comprehensive studies in larger cohorts, using the same treatment strategies, are needed to delineate the prognostic impact of concurrent HD and high-risk cytogenetics compared with that of NHD and high-risk cytogenetics. Assessment of HD by MC and iFISH may be mandatory for patients newly diagnosed as having MM.
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology (NRF-2012R1A1A2044138).
References
1. Rajkumar SV. Myeloma today: disease definitions and treatment advances. Am J Hematol. 2016; 91:90–100. PMID: 26565896.
2. Stella F, Pedrazzini E, Agazzoni M, Ballester O, Slavutsky I. Cytogenetic alterations in multiple myeloma: prognostic significance and the choice of frontline therapy. Cancer Invest. 2015; 33:496–504. PMID: 26506456.
3. Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. PMID: 19798094.
4. Carballo-Zarate AA, Medeiros LJ, Fang L, Shah JJ, Weber DM, Thomas SK, et al. Additional-structural-chromosomal aberrations are associated with inferior clinical outcome in patients with hyperdiploid multiple myeloma: a single-institution experience. Mod Pathol. 2017; 30:843–853. PMID: 28281554.
5. Hebraud B, Magrangeas F, Cleynen A, Lauwers-Cances V, Chretien ML, Hulin C, et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: the IFM experience. Blood. 2015; 125:2095–2100. PMID: 25636340.
6. Kumar S, Fonseca R, Ketterling RP, Dispenzieri A, Lacy MQ, Gertz MA, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012; 119:2100–2105. PMID: 22234687.
7. Merz M, Hielscher T, Seckinger A, Hose D, Mai EK, Raab MS, et al. Baseline characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am J Hematol. 2016; 91:E473–E477. PMID: 27508939.
8. Pawlyn C, Melchor L, Murison A, Wardell CP, Brioli A, Boyle EM, et al. Coexistent hyperdiploidy does not abrogate poor prognosis in myeloma with adverse cytogenetics and may precede IGH translocations. Blood. 2015; 125:831–840. PMID: 25428216.
9. International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003; 121:749–757. PMID: 12780789.
10. Shaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: an international system for human cytogenetic nomenclature (2013): Karger Medical and Scientific Publishers. 2013.
11. Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised International Staging System for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015; 33:2863–2869. PMID: 26240224.
12. Chretien ML, Corre J, Lauwers-Cances V, Magrangeas F, Cleynen A, Yon E, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015; 126:2713–2719. PMID: 26516228.
13. Lim JH, Seo EJ, Park CJ, Jang S, Chi HS, Suh C, et al. Cytogenetic classification in Korean multiple myeloma patients: prognostic significance of hyperdiploidy with 47-50 chromosomes and the number of structural abnormalities. Eur J Haematol. 2014; 92:313–320. PMID: 24372944.
14. Ortega M, Mallo M, Sole F, Sanchez-Morata C, Lopez-Andreoni L, Martinez-Morgado N, et al. 5q- syndrome and multiple myeloma diagnosed simultaneously and successful treated with lenalidomide. Leuk Res. 2013; 37:1248–1250. PMID: 23891188.
15. Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007; 110:719–726. PMID: 17420284.
16. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015; 33:3911–3920. PMID: 26282654.
17. Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci U S A. 2011; 108:632–637. PMID: 21187408.
18. Wirth M, Stojanovic N, Christian J, Paul MC, Stauber RH, Schmid RM, et al. MYC and EGR1 synergize to trigger tumor cell death by controlling NOXA and BIM transcription upon treatment with the proteasome inhibitor bortezomib. Nucleic Acids Res. 2014; 42:10433–10447. PMID: 25147211.
Supplementary Material
Supplemental Data Table S1
Characteristics of patients with multiple myeloma and hyperdiploidy, identified by a G-banded chromosome study and/or interphase FISH
Supplemental Data Table S2
Characteristics of patients with multiple myeloma and non-hyperdiploidy, identified by a G-banded chromosome study and/or interphase FISH
Fig. 1
Karyotype and interphase fluorescence in situ hybridization* using a D5S721/D5S23/EGR1 probe in multiple myeloma patients with chromosome 5 aberrations. (A) Case 9 showing partial trisomy 5; two normal chromosome 5s and one del(5q). (B) Case 38 showing del(5q). (C) Case 38 showing one red signal loss (2G1R), suggestive of del(5q). (D) Case 6 showing three green and red signals (3G3R), suggestive of trisomy 5 (left panel); and four green and three red signals (G4R3), suggestive of partial tetrasomy 5 (three normal chromosome 5s and one del(5q)) (right panel). (E) Case 16 showing G4R3, suggestive of partial tetrasomy 5. *FISH probe design is as follows: Chromosome 5p15 shows a green signal (G), and 5q31 shows a red signal (R). The normal FISH pattern of chromosome 5 is two green signals and two red signals (2G2R).
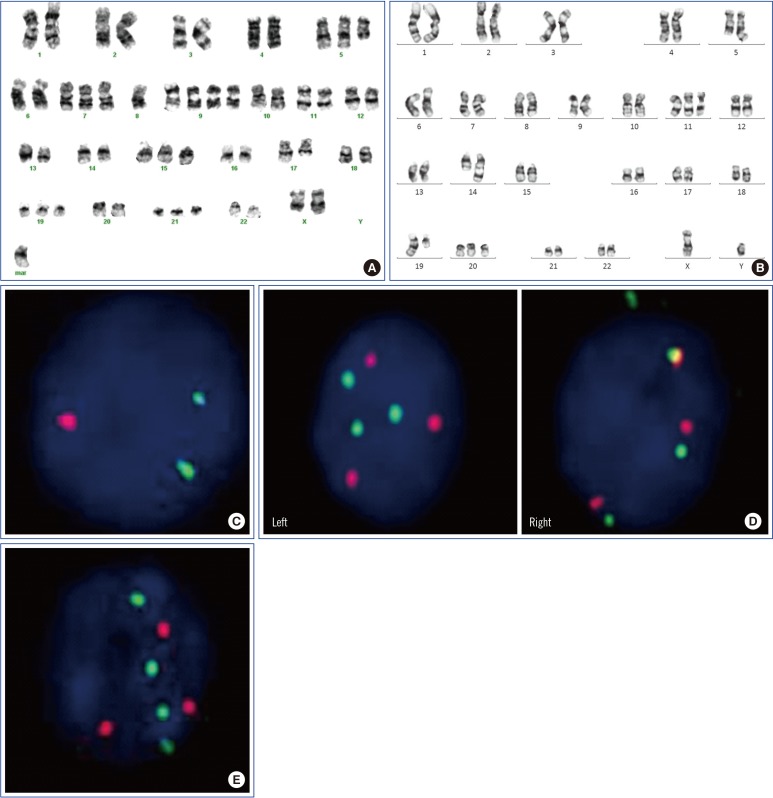
Table 1
Comparison of clinical and laboratory characteristics of patients with hyperdiploidy with high-risk cytogenetics and non-hyperdiploidy with high-risk cytogenetics
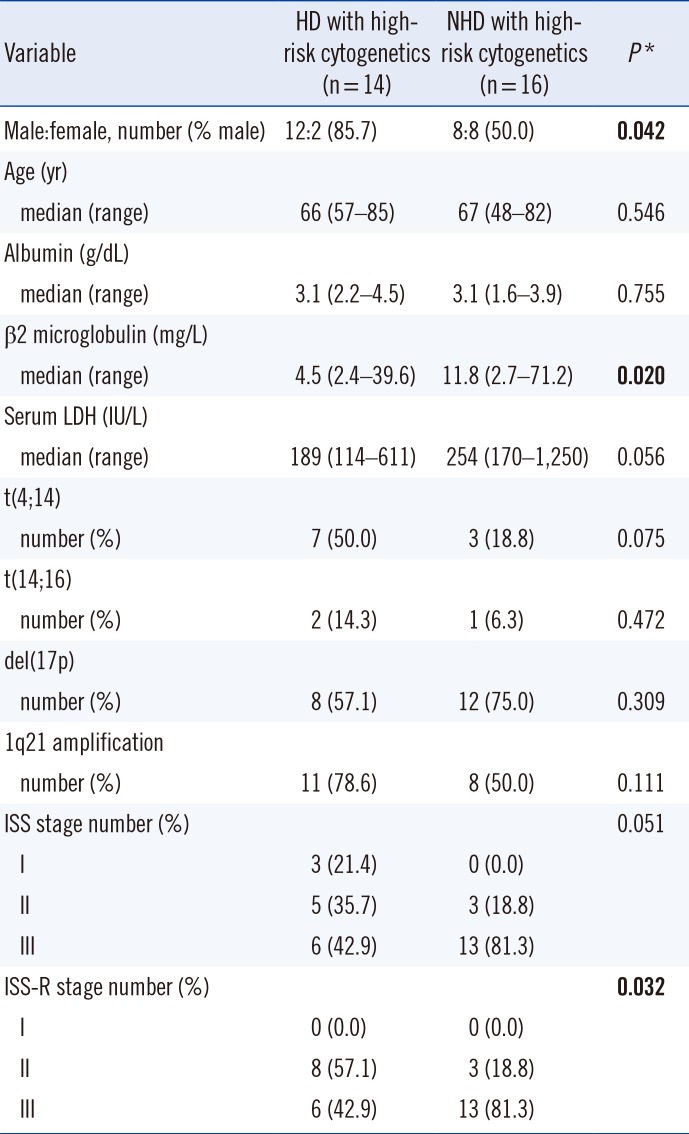
*P values were calculated by the χ2 test for categorical variables and the Mann-Whitney U test for continuous variables in a comparison between patients with HD with high-risk cytogenetics and those with NHD with high-risk cytogenetics. Significant values are shown in bold.
Abbreviations: HD, hyperdiploidy; ISS, International Staging System; ISS-R, Revised International Staging System; LDH, lactate dehydrogenase; NHD, non-hyperdiploidy.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download