Dear Editor,
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) is widely applied for the rapid identification of bacterial species [1]. However, an antimicrobial susceptibility test (AST) should be performed by using conventional methods following MALDI-TOF MS, and this delay represents a significant hurdle for the rapid adjustment of administered antimicrobial treatments. To overcome this limitation, many methods to improve MALDI-TOF MS for the production of rapid AST results have been proposed [234]. For example, carbapenemase-producing Enterobacteriaceae can be detected by analyzing the MALDI-TOF MS peaks of carbapenems and their metabolites [23].
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important multidrug-resistant organism in hospital and community settings. To date, few studies have attempted to differentiate MRSA and methicillin-susceptible S. aureus (MSSA) using MALDI-TOF MS. Despite the subsequent identification of many MRSA-specific peaks, no indisputable evidence has been obtained for reliable markers for MRSA and MSSA discrimination [567]. Furthermore, no study has evaluated Korean S. aureus isolates to date. Therefore, we aimed to identify MRSA isolates in Korea using MALDI-TOF MS peak analysis, and evaluated the feasibility of employing this method in clinical microbiology laboratories.
This study was approved by institutional review board of Yonsei university health system (Approval number 4-2017-0456) and informed consent was waived due to the retrospective manner of this study. We collected S. aureus isolated from the anterior nares of patients with atopic dermatitis from 2012 to 2013. Of 81 isolates, 47 were MSSA and 34 were MRSA as confirmed by the cefoxitin disc diffusion test according to the CLSI document M100-S25 [8]. All isolates were processed for protein extraction by using Microflex LT (Bruker Daltonics GmbH, Bremen, Germany) according to the manufacturer protocol. In Phase 1, peptide profiles were analyzed by using the QuickClassifier algorithm of ClinProTools 3.0 software (Bruker Daltonics GmbH). This model showed good sensitivity and specificity (Table 1). Using this high power of discrimination, we discovered peaks, which we called MRSA-specific peaks, at m/z 10,152 and 5,070. These peaks were detected in 27 of the 34 (79.4%) MRSA isolates and were completely absent in all MSSA isolates. To validate these findings, we isolated S. aureus from blood cultures referred in 2014 for Phase 2 of the study. Of the 92 S. aureus isolates, 54 were MRSA and 38 were MSSA. Although the same analysis was performed, the model showed a comparatively lower sensitivity and specificity than those from Phase 1 isolates (Table 1).
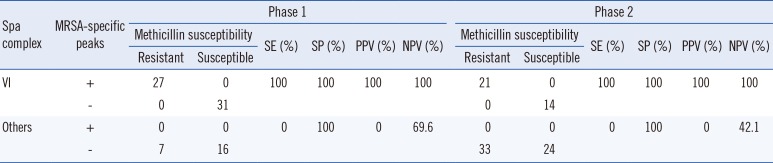
To evaluate whether the MRSA-specific peaks were associated with methicillin resistance in certain S. aureus clones, we determined the spa type of each S. aureus isolate as previously described [9]. The spa types of S. aureus isolates with MRSA-specific peaks showed repeat profiles corresponding with those of spa complex VI [10]. Of the 92 isolates belonging to spa complex VI, all MRSA isolates had MRSA-specific peaks (m/z 10,152 and 5,070), while the MSSA isolates did not (Table 2). To establish the composition of the MRSA-specific peaks, we processed the protein data using ProteinPilot (AB SCIEX, Foster City, CA, USA), but could not match proteins to the peaks.
Although several peptide spectra have been proposed to be useful for MRSA detection, the reliability of the MALDI-TOF MS result has been suggested to be problematic for this purpose. This controversy may come from differences in MALDI-TOF MS devices, matrix solutions, or even the different clones of each isolate among studies. To overcome these problems, we selected a widely used device and used protein extraction methods to obtain spectra with high resolution. However, our results demonstrated that discrimination of MRSA is indeed difficult when using MALDI-TOF MS spectra.
Although we failed to differentiate MRSA with comparable performance, our results show that a spa complex VI subset of MRSA could be correctly identified through the presence of MRSA-specific peaks. This finding supports the results of a previous study showing that MALDI-TOF MS-based identification of agr-positive MRSA was possible with high sensitivity and specificity [11]. In addition to the MRSA-specific peaks, we discovered peaks of m/z 10,100 and 5,050 that only existed when the MRSA-specific peaks were absent. Alterations in peptides involved in the mechanism of methicillin resistance are likely to explain this finding.
The differentiation of MRSA using MALDI-TOF MS is not currently feasible. Nevertheless, we found MRSA-specific peaks that allowed the rapid identification of certain spa-type MRSA isolates with high predictability. To our knowledge, this is the first report of MALDI-TOF MS protein peaks detected only in MRSA while absent in MSSA in certain spa-type S. aureus isolates. Further studies are required to establish the protein composition of these MRSA-specific peaks, preferred peptide preparation methods, and optimal analytic conditions.
References
1. Yu WS, Lee KM, Hwang KJ. Taxonomic identification of bacillus species using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Ann Clin Microbiol. 2016; 19:110–120.
2. Lasserre C, De Saint Martin L, Cuzon G, Bogaerts P, Lamar E, Glupczynski Y, et al. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol. 2015; 53:2163–2171. PMID: 25926485.
3. Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol. 2015; 53:1731–1735. PMID: 25694522.
4. Kim YA, Yong D, In YH, Park HS, Lee K. Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry to screen the extended-spectrum β-lactamase-producing ST131 Escherichia coli strains. Ann Clin Microbiol. 2016; 19:65–69.
5. Bernardo K, Pakulat N, Macht M, Krut O, Seifert H, Fleer S, et al. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics. 2002; 2:747–753. PMID: 12112858.
6. Du Z, Yang R, Guo Z, Song Y, Wang J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2002; 74:5487–5491. PMID: 12433077.
7. Szabados F, Kaase M, Anders A, Gatermann SG. Identical MALDI TOF MS-derived peak profiles in a pair of isogenic SCCmec-harboring and SCCmec-lacking strains of Staphylococcus aureus. J Infect. 2012; 65:400–405. PMID: 22750235.
8. Clinical and Laboratory Standards Institute. M100-S25. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute;2015.
9. Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999; 37:3556–3563. PMID: 10523551.
10. Ruppitsch W, Indra A, Stöger A, Mayer B, Stadlbauer S, Wewalka G, et al. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006; 44:2442–2448. PMID: 16825362.
11. Josten M, Dischinger J, Szekat C, Reif M, Al-Sabti N, Sahl HG, et al. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol. 2014; 304:1018–1023. PMID: 25116838.
Table 1
Sensitivity and specificity of MALDI-TOF MS using ClinProTool for differentiating MRSA isolates in Phase 1 and Phase 2
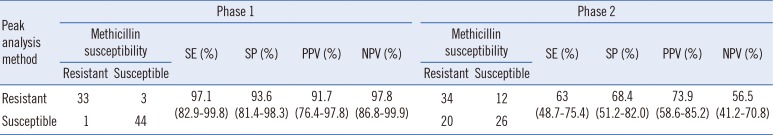
Table 2
Diagnostic capability of MRSA-specific peaks in spa complex VI and other complexes




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download