Abstract
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, with its accuracy and speed, is widely used for bacterial identification. The ASTA MicroIDSys system (ASTA, Korea) was recently developed for species identification. We compared its performance with that of Bruker Biotyper (Bruker Daltonics, Germany). Microbes were recovered from sputum, urine, and pus samples from patients admitted to a tertiary care hospital in Korea from January to April 2016. Matrix solution (α-cyano-4-hydroxycinnamic acid) was used, and the peptide profiles acquired from the Microflex LT (Bruker Daltonics) and Tinkerbell LT (ASTA) were analyzed by using their respective software. From 5,322 isolates, Bruker Biotyper identified 163 species; fifty species from 4,919 isolates were identified more than 10 times, including Klebsiella pneumoniae (n=571), Acinetobacter baumannii (n=436), Pseudomonas aeruginosa (n=358), Escherichia coli (n=372), Staphylococcus aureus (n=511), S. epidermidis (n=444), Enterococcus faecium (n=262), E. faecalis (n=220), and Candida albicans (n=248). Identical results, confidence scores (≥ 2.0 for Bruker Biotyper), and acceptable scores (≥140 for ASTA MicroIDSys) were obtained for 86.1% of isolates. Of 4,267 isolates, 99.2% showed acceptable scores in both systems. Results from the ASTA MicroIDSys showed good agreement with those from the Bruker Biotyper. The ASTA MicroIDSys could reliably identify clinically important microorganisms.
Bacterial identification with automated instruments or conventional methods such as biochemical reactions takes a few hours to days in clinical microbiology laboratories. More rapid methods are necessary to diagnose and treat septic patients, and better accuracy is necessary for classifying complicated bacterial mixtures. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is widely used for bacterial identification in clinical microbiology laboratories because of its speed and accuracy [123].
Two in vitro diagnostic MALDI-TOF MS systems, the Bruker Biotyper MS (Bruker Daltonics, Bremen, Germany) and the Vitek MS (bioMérieux, Marcy l'Etoile, France), have been implemented in clinical microbiology laboratories worldwide and are routinely used for identifying bacterial and yeast isolates [456]. Recently, a new system, the ASTA MicroIDSys system (ASTA, Suwon, Korea), was developed for identification of clinically important pathogenic species. The ASTA MicroIDSys system consists of a linear-type MALDI-TOF MS, a database, and software for species identification by spectral pattern matching. The linear-type MALDI-TOF MS performs microbial MS analysis in the range of m/z=2,000–20,000, with a mass accuracy and resolving power of 250 ppm and 1,000, respectively. The database contains reference MALDI spectra for 2,604 species. The MicroIDSys software employs an auto-selection algorithm for mass peaking of each species or strain of microorganism, in which the number of peaks that specifies each species is selected by the machine, based on pre-determined parameters, for better accuracy. The machine program itself selects parameters and masses as well as intensities of importance. In the present study, we compared the performance of the ASTA MicroIDSys system with that of the Bruker Biotyper MS system for identifying bacteria and yeast in routine clinical microbiology laboratory for the first time.
A total of 5,322 isolates were recovered from clinical specimens of urine, sputum, tracheal aspirate, wounds, and pus from patients admitted to a tertiary care hospital in Korea in January-April 2016. The specimens were inoculated in appropriate media such as 5% sheep blood agar, MacConkey agar, or chocolate agar for bacteria and Sabouraud dextrose agar for yeast, and then incubated for 24–48 hr at 35℃. A single bacterial colony from the agar was smeared onto the target plate (Bruker Daltonics GmbH), the matrix solution (α-cyano-4-hydroxycinnamic acid) was overlaid on the spot, and the peptide profile was acquired from the Bruker Microflex LT system. For yeast analysis, suspicious colonies were smeared directly onto the target plate and overlaid with 1 µL 70% formic acid (Sigma-Aldrich, St. Louis, MO, USA) and matrix solution. The Microflex system had the Biotyper software 3.1 and the MALDI Biotyper reference library version 5.0.0.0. The mass spectra were analyzed according to the manufacturer's instructions. We used identification score values ≥2.0 for bacteria and yeast. After complete analysis using the Bruker Biotyper, the peptide profiles were obtained by using the ASTA MicroIDSys on the same target plates. All the mass profiles were then analyzed by using the MicroIDSys 1.0. The cut-off value was set at ≥140 for the ASTA MicroIDSys for all microorganisms. PCR and 16S rRNA gene sequencing were performed for isolates that showed different results from the Bruker Biotyper and ASTA MicroIDSys systems.
Among the 5,322 isolates, 50 species (from 4,919 isolates) were isolated more than 10 times and analyzed for comparing the performances of the two MALDI-TOF MS systems. The results were as follows: 2,222 gram-negative bacilli, 1,926 gram-positive cocci, 413 Candida spp., and 385 other bacteria were detected. The most frequently isolated bacteria were Klebsiella pneumoniae (n=571), followed by Acinetobacter baumannii (n=436), Pseudomonas aeruginosa (n=358), Escherichia coli (n=372), Staphylococcus aureus (n=511), S. epidermidis (n=444), Enterococcus faecium (n=262), E. faecalis (n=220), Corynebacterium striatum (n=201), and Candida albicans (n=248).
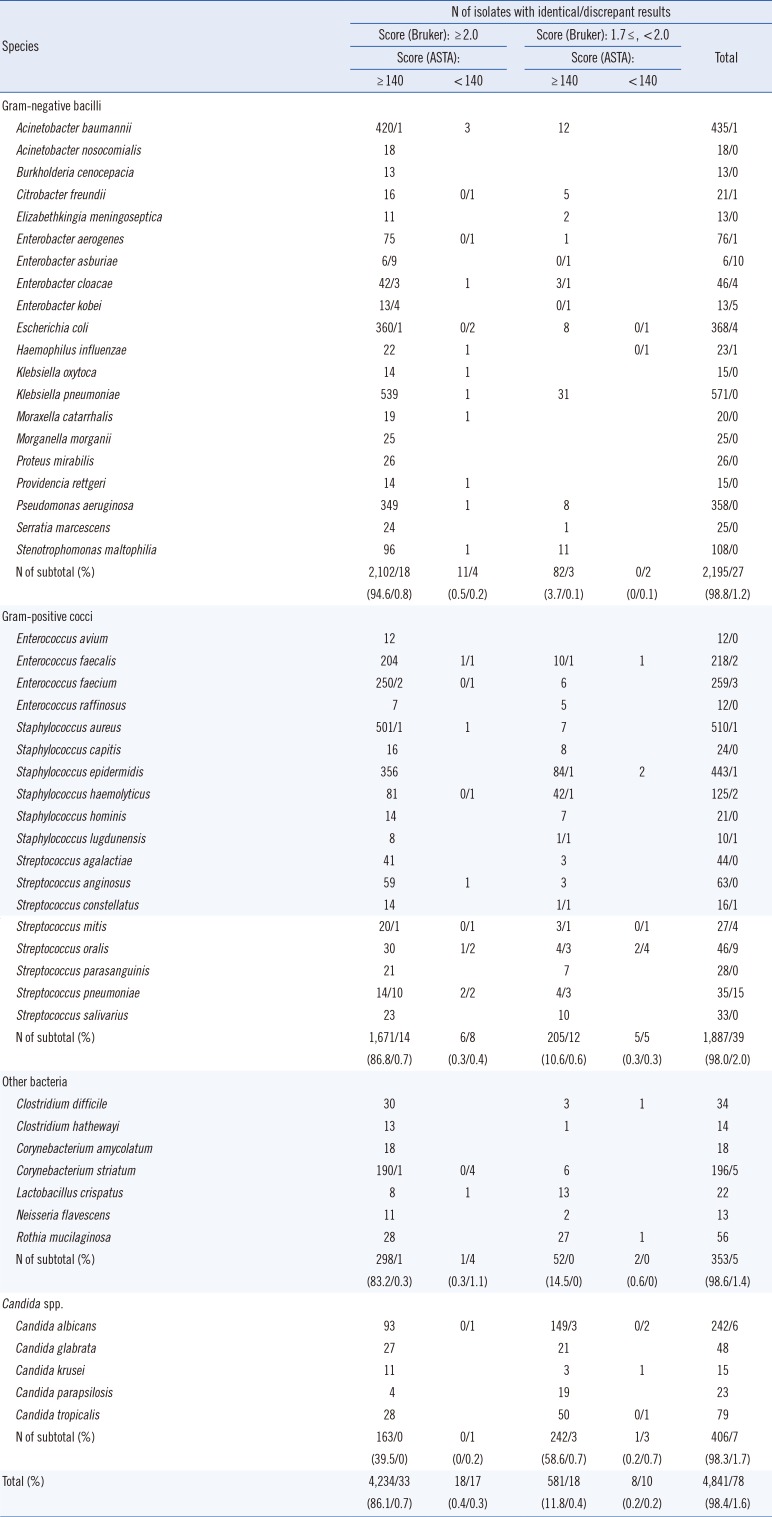
From the 4,919 isolates studied, identical results with confidence scores (≥2.0 for the Bruker Biotyper MS system) and acceptable scores (≥140 for the ASTA MicroIDSys system) were obtained for 4,234 (86.1%) isolates (Table 1). For the bacteria that are frequently isolated in clinical microbiology laboratories, the high agreement rates were as follows: K. pneumonia (100%), E. coli (98.9%), P. aeruginosa (100%), A. baumannii (99.8%), S. aureus (99.8%), S. epidermidis (99.8%), E. faecium (98.9%), and E. faecalis (99.1%). In addition, 4,841 (98.4%) isolates had a Bruker Biotyper score ≥1.7 and an ASTA MicroIDSys score ≥140. Only 78 (1.6%) isolates showed discrepant results between the two systems. For these isolates, we performed 16S rRNA gene sequencing. However, some species in the isolates were not accurately identified by either of the two methods; the 16S rRNA gene sequence similarity was very high for Enterobacter and Streptococcus mitis groups [7].
From the observed discrepant results between the two MALDI-TOF MS systems, we suspected that a known limitation of other MALDI-TOF MS systems might also be present in the ASTA MicroIDSys. Microorganisms are identified by MALDI-TOF MS systems using prerecorded protein spectra that are present in the system library, and these spectra are mostly based on ribosomal proteins. Therefore, MALDI-TOF MS systems are intrinsically limited to differentiate closely related species or strains of Salmonella spp., Raoutella, Klebsiella, Enterobacter, and Citrobacter [48].
Identical results, with scores between 1.7 and 2.0 for the Bruker Biotyper MS system and acceptable scores ≥140 for the ASTA MicroIDSys system, were obtained for 581 (11.8%) isolates; these included 242 (58.6%) Candida spp., 205 (10.6%) gram-positive cocci, and 82 (3.7%) gram-negative bacilli. Only two isolates of C. albicans showed discrepant results with an ASTA MicroIDSys score <140. This result suggested that the threshold score for identification with the Bruker Biotyper should be 1.7, instead of the usual 2.0, in order to compensate for the spectrum quality of Candida spp. In contrast, the cutoff score used for identification by the ASTA MicroIDSys was the typical 140 value itself, indicating that the power to discriminate between Candida species was higher in the ASTA MicroIDSys system than in the Bruker Biotyper system. The ASTA MicroIDSys MS system also showed high accuracy rates for overall identification of bacteria and Candida spp. from isolates.
In this study, we identified clinically relevant bacteria and Candida species from clinical specimens using the ASTA MicroIDSys system. Our findings on ASTA MicroIDSys system performance in the identification of bacteria and Candida species are in high agreement with findings from the Bruker Biotyper system. Especially for frequently isolated bacteria, such as K. pneumoniae, E. coli, P. aeruginosa, A. baumannii, S. aureus, S. epidermidis, E. faecium, and E. faecalis, high agreement rates (98.9–100%) were shown. In conclusion, the ASTA MicroIDSys has comparable identification capability to the Bruker Biotyper system. The ASTA MicroIDSys system can reliably identify microorganisms that are commonly isolated in clinical microbiological laboratories.
Acknowledgments
This work was supported by the National Research Foundation of Korea (2014M3A9E5073818), the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (H-GUARD_2014M3A6B2060509), and the Ministry of Health & Welfare, Republic of Korea (HI14C1324).
We are deeply grateful to Yong Ha In, Kyu Hwan Park, and Hyung Soon Park (ASTA Inc.), as well as to Hyungsun Kim and Sori Jong (The Research Institute of Antimicrobial Resistance, Yonsei University College of Medicine) for their help.
References
1. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009; 49:543–551. PMID: 19583519.
2. Claydon MA, Davey SN, Edwards-Jones V, Gordon DB. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol. 1996; 14:1584–1586. PMID: 9634826.
3. Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012; 50:1313–1325. PMID: 22322345.
4. Bilecen K, Yaman G, Ciftci U, Laleli YR. Performances and reliability of Bruker Microflex LT and VITEK MS MALDI-TOF mass spectrometry systems for the identification of clinical microorganisms. Biomed Res Int. 2015; 2015:516410. PMID: 26793718.
5. Richter SS, Sercia L, Branda JA, Burnham CA, Bythrow M, Ferraro MJ, et al. Identification of Enterobacteriaceae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using the VITEK MS system. Eur J Clin Microbiol Infect Dis. 2013; 32:1571–1578. PMID: 23818163.
6. Bessède E, Angla-Gre M, Delagarde Y, Sep Hieng S, Ménard A, Mégraud F. Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a university hospital. Clin Microbiol Infect. 2011; 17:533–538. PMID: 20518792.
7. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007; 45:2761–2764. PMID: 17626177.
8. Wang H, Fan YY, Kudinha T, Xu ZP, Xiao M, Zhang L, et al. A comprehensive evaluation of the Bruker Biotyper MS and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of yeasts, part of the national china hospital invasive fungal surveillance net (CHIF-NET) study, 2012 to 2013. J Clin Microbiol. 2016; 54:1376–1380. PMID: 26912761.
SUPPLEMENTARY MATERIAL
Supplemental Data Table S1
Comparison of the identified bacteria and yeast with score values ≥2.0 for the Bruker Biotyper system and ≥140 for the ASTA MicroIDSys system
Table 1
Comparison of the results for frequently isolated bacteria from the Bruker Biotyper and ASTA MicroIDSys systems




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download