Abstract
Background
Cases of infective endocarditis (IE) require prompt etiological diagnosis for effective treatment. Molecular methods can aid in rapid and reliable diagnosis of culture-negative IE cases. We evaluated the utility of 16S rDNA PCR and sequencing in determining the causative agents of IE in valve tissues, especially when specimens were obtained after initiation of antimicrobial therapy.
Methods
We performed 16S rDNA PCR and sequencing in heart valve specimens and medical records review of 80 patients who underwent protocol-based cardiac surgery from 2013 to 2015. One patient did not meet the criteria for IE. Sixty-five (81.3%) and 14 pa-tients (17.5%) were diagnosed as having definite IE and possible IE, respectively. Blood and heart valve biopsy tissue were examined by using routine microbiological methods.
Results
Blood cultures in our hospital were IE-positive for 33 patients (41.8%), whereas 49 patients (62.0%) showed positive blood cultures when initial blood cultures performed at the referring hospital were included. Eighteen (22.8%) and 40 patients (50.6%) were IE-positive in valve tissue cultures and 16S rDNA PCR, respectively. Bacteria in the Streptococcus mitis group (n=26) were the most common etiological agents of IE. Eight (10.1%) culture-negative specimens tested positive by 16S rDNA PCR. In five of eight PCR-positive and culture-negative cases, fastidious or anaerobic organisms were the cause of IE.
Infective endocarditis (IE) is a serious infectious disease that requires early detection [1]. The modified Duke criteria for diag-nosis of IE include the patient's condition, microbiological evidence, and medical imaging, including echocardiography. Blood culture and heart valve biopsy culture are the key confirmatory tests used for clinical diagnosis of IE. However, the two tests often do not detect rare and/or difficult-to-culture causative agents. About 5% (range, 2.5–31%) of all IE cases are associated with negative blood cultures [2], which renders prescription of specific antibiotics to these patients difficult [3]. Since April 2013, we have applied PCR for amplifying and sequencing the gene encoding the 16S rRNA (16S rDNA) in heart valve specimens to detect and identify bacterial DNA [456]. PCR has assisted in correctly diagnosing the blood culture-negative cases of IE, including those caused by fastidious or non-cultivable microbes [789].
We evaluated the utility of 16S rDNA PCR amplification from heart valve tissues for the etiological diagnosis of patients undergoing cardiac surgery as a result of IE, especially when specimens were obtained after initiation of antimicrobial therapy.
From April 2013 to September 2015, patients with suspected IE who were admitted and underwent valve replacement surgery were enrolled prospectively. Patients with definite or possible IE were diagnosed by using the modified Duke criteria [1011]. The modified Duke criteria consist of three categories: two pathological criteria, two major clinical criteria, and five minor clinical criteria. Cases of definite IE were defined to include cases with one or more pathological criteria, two major clinical criteria, one major and three minor clinical criteria or five minor clinical criteria. Possible patients with IE were defined as those having one major and one minor clinical criterion or three minor clinical criteria. The patients' medical records were reviewed, and clinical information was collected, including demographics, underlying conditions, previous antibiotic treatment, and laboratory findings.
Of the 80 patients enrolled in this study, only one patient was determined to be IE-negative. Sixty-five (82.3%) and 14 patients (17.7%) were diagnosed as having definite IE and possible IE, respectively. Table 1 shows the demographic characteristics and clinical features of the 79 patients with IE. Twenty patients (25.3%) had been treated for cardiovascular disease or endocarditis. Native valve endocarditis accounted for 69 (87.3%) IE cases. Four patients had dental problems, and two patients had undergone dental procedures. All patients were treated with antibiotics before valve surgery. The median duration (and range) of pre-operative antibiotic treatment was 5 (1–150) days. The median duration of pre-operative antibiotic treatment for 17 patients who were positive by PCR and by valve culture was 1 (1–17) days. The median duration of pre-operative antibiotic treatment for 26 patients who were positive by PCR but negative by valve culture was 6 (1–58) days.
The study was approved by the Institutional Review Board of the Asan Medical Center, Seoul, Korea; each patient provided a written informed consent before enrollment.
Three sets of blood culture were obtained for each patient with suspected IE. A continuous monitoring blood culture system, BACTEC FX system (Becton-Dickinson, Franklin Lakes, NJ, USA), was used to culture blood with BACTEC Plus Aerobic/F medium and BACTEC Lytic/10 Anaerobic/F medium (Becton-Dickinson). Specimens that generated positive signal in the BACTEC culture system were tested with MicroScan Combo panels (Siemens Healthcare Systems, Malvern, PA, USA), MALDI Biotyper (Bruker Daltonics, Billerica, MA, USA) matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) system, VITEK2 NH (BioMérieux, Marcy-l'Étoile, France), and/or ANC cards to identify the causative pathogens. The valve specimens were homogenized by bead beating using the FastPrep-24 instrument (MP Biomedicals, Santa Ana, CA, USA). The disrupted specimens were divided into two parts: one was used for culture, and the other was directly processed and used for 16S rDNA PCR. The disrupted valve specimens were cultured on blood agar plate and Brucella agar plate, and in thioglycolate broth. The positive tissue cultures were identified by using the same methods as the positive blood cultures.
DNA was extracted from the disrupted specimen by using a QIA-amp DNA mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. DNA was eluted in a final volume of 50 µL and 5 µL DNA template was used for each PCR reaction. 16S rDNA PCR was used to amplify the 16S rDNA. The following two primer sets were used: 8FPL, 5′-AGT TTG ATC CTG GCT CAG-3′, and 806R, 5′-GGA CTA CCA GGG TAT CTA AT-3′; 515FPL, 5′-TGC CAG CAG CCG CGG TAA-3′, and 13B, 5′-AGG CCC GGG AAC GTA TTC AC-3′ [1213]. Each run included positive and negative controls. A Dyne PCR Purification Kit (DYNEBIO Inc., Seongnam, Korea) was used to purify amplicons. Direct sequencing of amplicons was carried out by analysis of the amplified products by capillary electrophoresis using the ABI PRISM 3730xl genetic analyzer (Applied Biosystems). Sequencing reactions were prepared according to the manufacturer's instructions (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit; Applied Biosystems). Guideline MM18-A of the Clinical and Laboratory Standards Institute was applied for quality control of sequences [14].
We used the GenBank database and EzTaxon-e [15] to compare the sequences obtained with those of reference organisms [16]. Bacteria were identified according to the criteria specified in the CLSI MM18-A guideline [14]. Descriptive statistics were used to describe the basic features of the data. The median durations of antibiotic use were compared by using the Mann-Whitney U test of SPSS 21.0 (IBM Corporation, Armonk, NY, USA).
Blood cultures in our hospital were positive for 33 patients (41.8%). Forty-nine patients (62.0%) had positive blood cultures when combined with the results of the initial blood cultures performed at the referring hospital. Fig. 1 shows a comparison of the results of blood culture, biopsy culture, and/or 16S rDNA PCR. Fifty-nine cases showed at least one positive result from the three microbiological tests. Since six patients met the clinical criteria (Dukes modified criteria) for IE, they were diagnosed as having definite IE without microbiological confirmation. Twenty (25.3%) and 43 (54.4%) patients were positive for IE in valve tissue cultures and 16S rDNA PCR, respectively. Fifteen cases showed positive results by all three tests.
The most common etiological organisms were bacteria in the Streptococcus mitis group (n=26), followed by Staphylococcus aureus (n=10), Enterococcus spp. (n=4), Staphylococcus epidermidis (n=3), and nutritionally variant streptococci (n=3) (Table 2). Eight cases were positive by 16S rDNA PCR only, and the following bacterial etiologies were identified: Streptococcus constellatus, Streptococcus sanguinis, Streptococcus salivarius, Haemophilus parainfluenzae, Granulicatella adiacens, Aggregatibacter actinomycetemcomitans, Abiotrophia spp., and Propionibacterium humerusii. Three cases were positive for both culture methods, but negative for 16S rDNA PCR. The organisms present in these cases were S. aureus, S. epidermidis, and Enterococcus faecium. Two cases that were blood culture-negative were positive by 16S rDNA PCR and biopsy culture of heart valve specimens; S. mitis group and Streptococcus agalactiae were determined as the causes of IE in these cases.
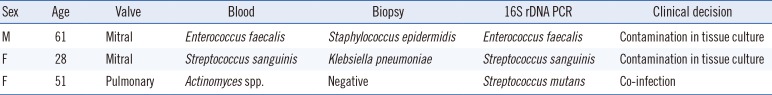
Discrepancies between blood culture, heart valve biopsy culture, and 16S rDNA PCR and sequencing results were observed in three patients with definite IE (Table 3). In the case of a 61-yr-old man, both the blood culture and 16S rDNA PCR indicated infection with Enterococcus faecalis, whereas the heart valve culture was positive for S. epidermidis. Similarly, in the case of a 28-yr-old woman, blood culture and 16S rDNA PCR results indicated S. sanguinis infection, but the biopsy culture was positive for Klebsiella pneumoniae. The heart valve specimen of a 51-old-yr woman indicated infection with different organisms by blood culture (Actinomyces spp.) and 16S rDNA PCR and sequencing (Streptococcus mutans). This was determined as a case of co-infection.
We determined the microbiological etiologies of 79 patients with IE who received protocol-based cardiac surgery for 2 yr and 5 months. Blood culture remains an essential method of detecting IE. The proportion of blood culture-negative cases was 38.0%, which is higher than that reported in previous studies (7–20%) [17]. Since some patients were not diagnosed as having IE initially and had received empirical antibiotics, this high proportion of blood culture-negative cases might have resulted from previous antibiotic use.
The culture-positive valve specimens were 39.5% (17/43) among patients who tested positive by 16S rDNA PCR. In previous studies, the percentage of agreement between positive PCR and valve culture results was 11.1–93.1% [18]. Our study showed a slightly higher percentage of cases that were positive by both valve culture and 16S rDNA PCR. This might be associated with the duration of pre-operative antibiotic use. The average duration of pre-operative antibiotic use in this study was less than that in previous studies [19]. In a previous study, the median duration of pre-operative antibiotic use was 4 (2–8) days for patients whose valve specimens were positive by both PCR and valve culture and 15.5 (1–58) days for patients whose valve specimens were positive by PCR but negative by culture [19]. Our data showed that bacterial DNA might persist during antibiotic treatment in infected valve tissue for long periods.
Direct 16S rDNA PCR is useful for identifying the etiologies of IE in blood culture- or biopsy culture-negative cases, especially with fastidious organisms such as Brucella spp., Coxiella burnetii, Bartonella spp., Tropheryma whipplei, Mycoplasma spp., and Legionella spp. [20]. In the present study, five out of eight culture-negative/16S rDNA PCR-positive cases were caused by fastidious or anaerobic organisms. The application of broad-spectrum PCR significantly increases the ability to detect difficult-to-culture organisms, including dead bacteria [2122]. Three cases were IE-positive by both culture methods, but negative by 16S rDNA PCR. The organisms present in these cases were not classified as fastidious organisms. PCR assays are sensitive but not without limitations, which include the presence of PCR inhibitors in clinical samples, as well as the risk of contamination in clinical samples and PCR reagents. In addition, biopsy samples may not be homogenous, and the results may reflect molecular assays processed from sterile sites.
Of the three cases which showed discrepancies in the results of the three tests, two cases were positive for the same organ-ism by blood culture and 16S rDNA PCR, but the biopsy culture was positive for different organism. One of those, the biopsy culture was positive for K. pneumoniae, which was multi-drug resistant and isolated only in enrichment medium, suggesting possible contamination during inoculation [23]. Since the positive heart valve culture for the different organism was interpreted as a contaminant, 16S rDNA PCR and sequencing was helpful in identifying contaminated culture. In the other case, 16S rDNA PCR and blood culture identified different pathogens, and the attending physician prescribed antibiotics to treat both organisms. Thus, 16S rDNA PCR may be a useful tool to complement conventional culture methods.
Our study has several limitations. First, we did not clone bacterial DNA; therefore, cases of polymicrobial infections could not be detected. This might have resulted in the relatively low number of 16S rDNA PCR-positive specimens in our study. Second, we did not include a control group. We could not evaluate the specificity, positive predictive value, or negative predictive value of the 16S rDNA PCR as almost all the patients had IE.
In conclusion, 16S rDNA PCR with sequencing is a useful supportive tool for the etiological diagnosis of IE, especially for cases of culture-negative IE. PCR and sequencing assays are particularly useful in the diagnosis of IE in patients who have had prior antimicrobial therapy, had tested positive by blood culture owing to suspected contaminants, or are predisposed towards rare infections such as Bartonella endocarditis owing to homelessness and alcoholism [3].
Acknowledgments
This study was supported by the BioNano Health-Guard Research Center, funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontiers Project (Grant Number H-GUARD_ERND2-5).
References
1. Wouters BJ, Sanders MA, Lugthart S, Geertsma-Kleinekoort WM, van Drunen E, Beverloo HB, et al. Segmental uniparental disomy as a recurrent mechanism for homozygous CEBPA mutations in acute myeloid leukemia. Leukemia. 2007; 21:2382–2384. PMID: 17554374.
2. Lamas CC, Eykyn SJ. Blood culture negative endocarditis: analysis of 63 cases presenting over 25 years. Heart. 2003; 89:258–262. PMID: 12591823.
3. Gould FK, Denning DW, Elliott TS, Foweraker J, Perry JD, Prendergast BD, et al. Guidelines for the diagnosis and antibiotic treatment of endocarditis in adults: a report of the Working Party of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother. 2012; 67:269–289. PMID: 22086858.
4. Nikkari S, Merilahti-Palo R, Saario R, Söderström KO, Granfors K, Skurnik M, et al. Yersinia-triggered reactive arthritis. Use of polymerase chain reaction and immunocytochemical staining in the detection of bacterial components from synovial specimens. Arthritis Rheum. 1992; 35:682–687. PMID: 1599522.
5. Kotilainen P, Jalava J, Meurman O, Lehtonen OP, Rintala E, Seppälä OP, et al. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998; 36:2205–2209. PMID: 9665992.
6. Rantakokko-Jalava K, Nikkari S, Jalava J, Eerola E, Skurnik M, Meurman O, et al. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000; 38:32–39. PMID: 10618059.
7. Greub G, Lepidi H, Rovery C, Casalta JP, Habib G, Collard F, et al. Diagnosis of infectious endocarditis in patients undergoing valve surgery. Am J Med. 2005; 118:230–238. PMID: 15745720.
8. Rovery C, Greub G, Lepidi H, Casalta JP, Habib G, Collart F, et al. PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J Clin Microbiol. 2005; 43:163–167. PMID: 15634966.
9. Houpikian P, Raoult D. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore). 2005; 84:162–173. PMID: 15879906.
10. Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG Jr, Ryan T, et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis. 2000; 30:633–638. PMID: 10770721.
11. Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med. 1994; 96:200–209. PMID: 8154507.
12. Relman DA. Universal bacterial 16S rDNA amplification and sequencing. Diagnostic molecular microbiology: principles and applications. Washington DC: American Society for Microbiology;1993. p. 489–495.
13. Choi SH, Sung H, Kim SH, Lee SO, Lee SH, Kim YS, et al. Usefulness of a direct 16S rRNA gene PCR assay of percutaneous biopsies or aspirates for etiological diagnosis of vertebral osteomyelitis. Diagn Microbiol Infect Dis. 2014; 78:75–78. PMID: 24231384.
14. CLSI. Interpretive criteria for identification of bacteria and fungi by dna target sequencing; Approved guideline MM18-A. Wayne, PA: CLSI;2008.
15. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–721. PMID: 22140171.
16. Park KS, Ki CS, Kang CI, Kim YJ, Chung DR, Peck KR, et al. Evaluation of the GenBank, EzTaxon, and BIBI services for molecular identification of clinical blood culture isolates that were unidentifiable or misidentified by conventional methods. J Clin Microbiol. 2012; 50:1792–1795. PMID: 22403421.
17. Pant S, Patel NJ, Deshmukh A, Golwala H, Patel N, Badheka A, et al. Trends in infective endocarditis incidence, microbiology, and valve replacement in the United States from 2000 to 2011. J Am Coll Cardiol. 2015; 65:2070–2076. PMID: 25975469.
18. Miller RJ, Chow B, Pillai D, Church D. Development and evaluation of a novel fast broad-range 16S ribosomal DNA PCR and sequencing assay for diagnosis of bacterial infective endocarditis: multi-year experience in a large Canadian healthcare zone and a literature review. BMC Infect Dis. 2016; 16:146. PMID: 27066823.
19. Kotilainen P, Heiro M, Jalava J, Rantakokko V, Nikoskelainen J, Nikkari S, et al. Aetiological diagnosis of infective endocarditis by direct amplification of rRNA genes from surgically removed valve tissue. An 11-year experience in a Finnish teaching hospital. Ann Med. 2006; 38:263–273. PMID: 16754257.
20. Fournier PE, Watt G, et al. Blood culture-negative endocarditis. In : Habib G, editor. Infective endocarditis: epidemiology, diagnosis, imaging, therapy, and prevention. 1st ed. Switzerland: Springer International Publishing AG;2016. p. 245–258.
21. Horstkotte D, Follath F, Gutschik E, Lengyel M, Oto A, Pavie A, et al. Guidelines on prevention, diagnosis and treatment of infective endocarditis executive summary; the task force on infective endocarditis of the European society of cardiology. Eur Heart J. 2004; 25:267–276. PMID: 14972429.
22. Task Force sull'ndocardite Infettiva della Societá Europea di Cardiologia. Guidelines on prevention, diagnosis and treatment of infective endocarditis. Ital Heart J Suppl. 2004; 5:548–590. PMID: 15490689.
23. Morris AJ, Wilson SJ, Marx CE, Wilson ML, Mirrett S, Reller LB. Clinical impact of bacteria and fungi recovered only from broth cultures. J Clin Microbiol. 1995; 33:161–165. PMID: 7699035.
Fig. 1
Comparison of blood culture, heart valve or vegetation biopsy culture, and/or 16S rDNA PCR results in cases treated for IE. The case numbers indicate positive results by blood culture, biopsy culture, and 16S rDNA PCR results (n=49, 20, and 43, respectively). Twenty cases were negative by all three methods.
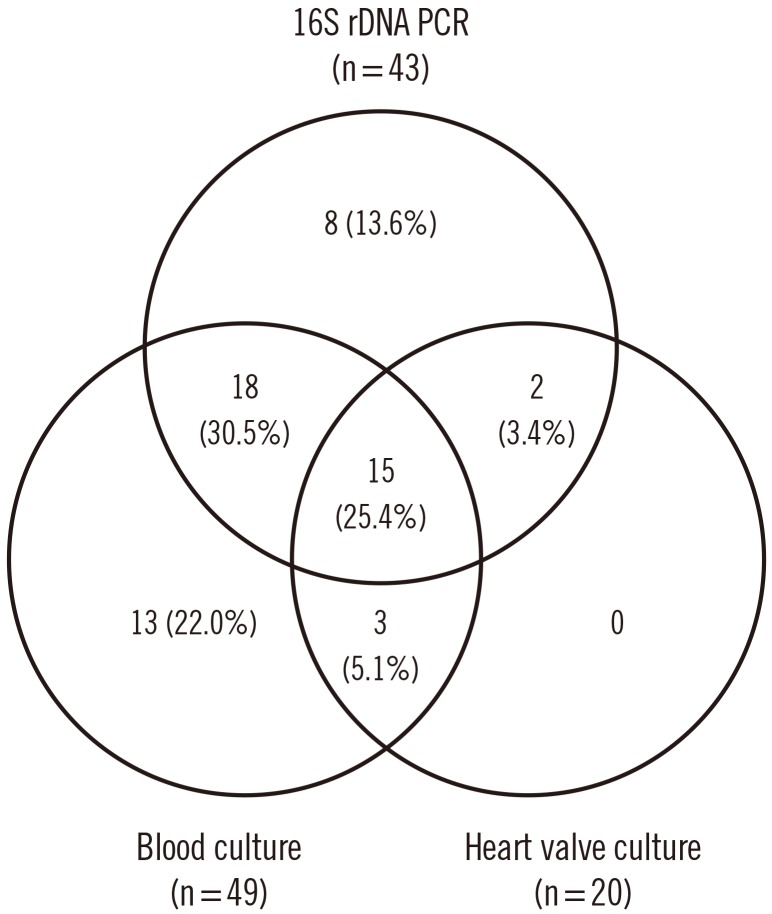
Table 1
Clinical characteristics of patients with definite and possible infective endocarditis (IE) (n=79)
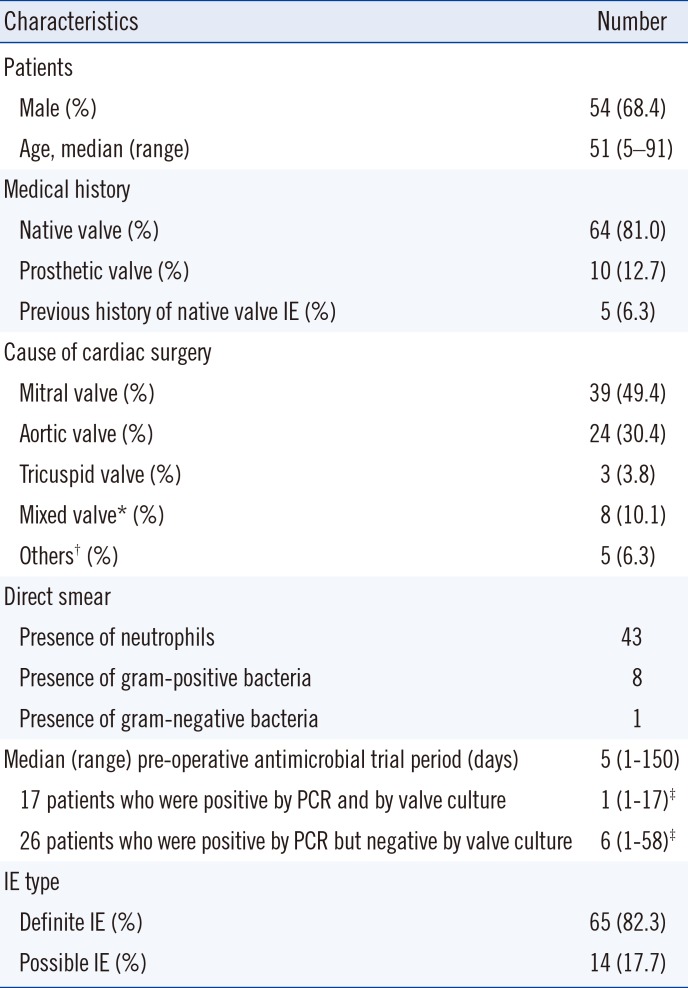
Table 2
Comparison of blood culture, vegetation biopsy culture, and 16S rDNA PCR results
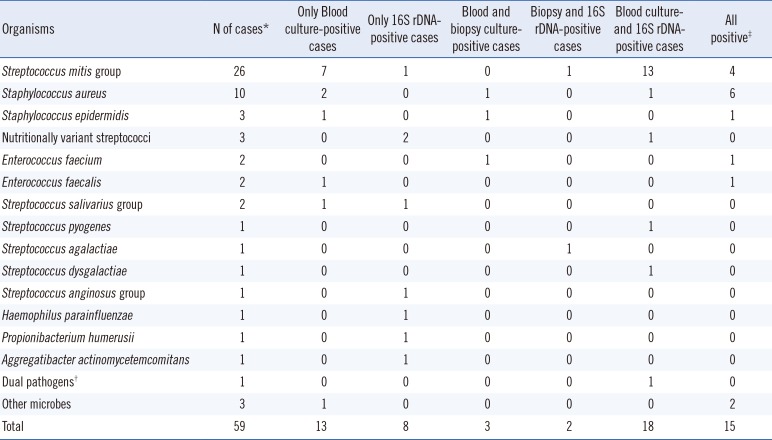
*Cases with positive result by any of the three methods (blood culture, vegetation biopsy culture, and/or 16S rDNA PCR); †Two different organisms identified by blood culture (Actinomyces spp.) and 16S rDNA PCR (S. mutans group); ‡Two cases with vegetation biopsy culture contaminations were excluded.
Table 3
Definite IE cases with discrepancies in results by blood culture, vegetation biopsy culture, and 16S rDNA PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download