Abstract
Background
Forkhead box P3 (Foxp3) is the most reliable marker for regulatory T cells, which play an important role in maintaining renal allograft tolerance. Recently, Foxp3 polymorphisms have been reported to be associated with graft outcome in kidney transplantation. We analyzed the association of Foxp3 polymorphisms with renal allograft outcome.
Methods
Foxp3 polymorphisms (rs3761548 A/C, rs2280883 C/T, rs5902434 del/ATT, and rs2232365 A/G) were tested by PCR with sequence-specific primers (PCR-SSP) in 231 adult kidney transplantation recipients from 1996-2004 at Seoul National University Hospital.
Results
Patients with the rs3761548 CC genotype showed better graft survival than those with the AC or AA genotype (log rank test, P=0.03). Patients with the rs3761548 CC genotype also showed a lower rate of recurrence of the original glomerular disease than those with the AC or AA genotype (P=0.01). The frequency of acute rejection (AR) in patients with the rs2280883 TT genotype was lower than that in patients with the rs2280883 CT or CC genotype (26.9% vs 53.3%, P=0.038). Patients with the rs2280883 TT genotype also showed better graft survival than those with the CT or CC genotype (P=0.03).
Improved pre-transplantation evaluation and the development of post-transplantation immunosuppressive therapy have led to a marked improvement in short-term graft survival in kidney transplantation. However, long-term graft survival remains unsatisfactory [1]. Immunologic responses of patients play pivotal roles in graft rejection or recurrence of underlying renal disease. Regulatory T cells (Tregs) promote a state of antigen-specific peripheral tolerance by suppressing activation and expansion of T effector cells, as reported in experimental models [23]; therefore, they play an important role in maintaining self-tolerance and in regulating graft rejection and graft-versus-host disease [45].
Forkhead box P3 (Foxp3) is a transcription factor that regulates Treg development and function and remains the most reliable marker for Treg [6]. Therefore, Foxp3 polymorphisms, which could affect the function and quantity of the Foxp3 molecule and thus result in Treg function defects, have been associated with various autoimmune diseases [78].
For kidney transplantation, the impact of Foxp3+ Tregs on graft outcomes seems conflicting in previous reports [9101112131415]. In some study, the presence of intragraft Tregs have been associated with favorable renal allograft outcome [910]. The FoxP3+ Treg/CD3+ T cell ratio positively correlated with graft function at two years after transplantation [9]. These cells could direct a FoxP3-induced immune response toward suppression of T effector cells, promoting renal graft acceptance with improved function. Lower level of intragraft Foxp3 mRNA predicts progression in renal transplants with borderline change [11]. The mRNA levels of Foxp3 in peripheral blood were higher in patients with operational tolerance or stable kidney graft function compared with patients with chronic rejection [1213]. However, other groups reported that mRNA for Foxp3 in the urine of recipients with acute rejection (AR) was higher than recipients with normal biopsy [14] and association of higher density of FoxP3+ cells with worse graft outcome in recipients with acute cellular rejection [15].
Recently, an association between Foxp3 polymorphisms and graft outcome has been reported also with conflicting results [161718]. Therefore, we analyzed the association of four Foxp3 single nucleotide polymorphisms (SNPs) (rs3761548 A/C, rs2280883 C/T, rs5902434 del/ATT, and rs2232365 A/G) with graft outcome in kidney transplantation.
This study included 231 kidney transplantation cases performed between January 1996 and December 2004 at the Seoul National University Hospital, Seoul, Korea. The baseline characteristics of the 231 kidney transplant recipients are shown in Table 1. Residual DNA samples were collected after routine preoperative tests for HLA genotype. DNA samples from 195 healthy Korean individuals studied in our previous cohort were used as the normal controls [19]. Samples were preserved at –70℃ prior to the experiments performed for this study. The following recipient characteristics were collected retrospectively: age and gender of recipient; age and gender of donor; type of donor (living vs cadaveric donor); primary renal disease causing end-stage renal disease; number of HLA mismatches; number of HLA-DR mismatches; crossmatch result at the time of transplantation; duration of hemodialysis; type of immunosuppression; time of transplantation; occurrence and time point of biopsy-proven AR; recurrence and time point of primary renal disease; 1-, 3-, 5-, and 10-yr creatinine levels post-transplantation; and occurrence and time point of graft failure, defined as graft nephrectomy or return to hemodialysis. The study protocol was designed in accordance with the Declaration of Helsinki and approved by the institutional review board of Seoul National University Hospital (IRB No. 1306-121-501).
DNA was extracted from the peripheral blood of 231 patients who had kidney transplantation between January 1996 and December 2004 by using the LaboPass Genomic DNA Extraction Kit (COSMO, Seoul, Korea) or QuickGene DNA whole blood kit (Fujifilm, Tokyo, Japan) when HLA genotyping for pre-operation evaluation was performed. For 195 healthy controls, DNA was extracted using LaboPass Genomic DNA Extraction Kit (COSMO, Seoul, Korea) during the period of January 1999 and July 2001. All DNA samples were preserved at –70℃ prior to being used for Foxp3 polymorphisms analyses which were performed during the period of June 2015 and July 2016. Four Foxp3 polymorphisms (rs3761548 A/C, rs2280883 C/T, rs5902434 del/ATT, and rs2232365 A/G) were analyzed by PCR with sequence-specific primers (PCR-SSP) with some modifications [20] (Table 2). PCR was performed by using a 40-µL reaction mixture containing 40 ng DNA, 0.2mM of each primer, 0.8 µL of 10mM dNTP, 2.0mM MgCl2, 1.0 U Taq DNA polymerase (Roche Applied Science, Basel, Switzerland), and 4 µL of 10× reaction buffer. The PCR protocol consisted of an initial denaturation step at 95℃ for 5 min; 35 cycles of denaturation at 95℃ for 30 sec, annealing (temperatures detailed in Table 2) for 30 sec, and extension at 72℃ for 30 sec; and a final extension step at 72℃ for 5 min.
Differences of allele frequency and genotype frequency were compared by using a 2-sided Chi-square test or Fisher's exact test, as appropriate. The odds ratio (OR) was calculated by using a 95% confidence interval (CI). Potential associations of variables with graft survival were analyzed by using the Cox proportional hazards regression model. Death-censored graft survival was analyzed by using the Kaplan-Meier method and the log rank test. A P value of <0.05 was considered statistically significant. SPSS for Windows version 18.0 (SPSS, Chicago, IL, USA) was used for statistical analysis.
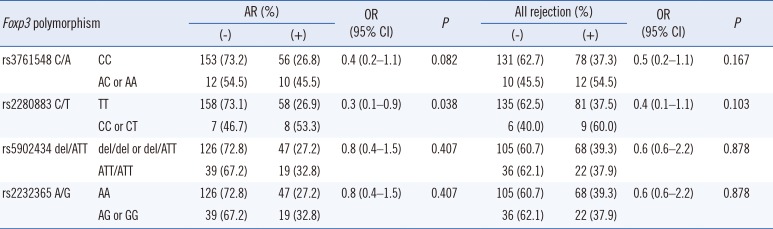
The frequencies of four Foxp3 SNPs in kidney transplant recipients were not statistically different from those of the normal controls (data not shown). Univariate Cox regression analysis indicated that the Foxp3 rs3761548 AC or AA genotype, rs2280883 CT or CC genotype, and AR episode were associated with graft survival (P=0.038, P=0.032, and P<0.001, respectively; Table 2).
The 1-, 3-, 5-, and 10-yr creatinine levels post-transplantation did not differ according to the Foxp3 genotypes (data not shown). However, AR frequency was lower in patients with the rs2280883 TT genotype than in patients with the CT or CC genotype (26.9% vs 53.3%, P=0.035, OR=0.3, 95% CI 0.1–0.9; Table 3). Kaplan-Meier survival analysis indicated that patients with the rs3761548 CC genotype (n=209) showed better graft survival than those with the AC or AA genotype (n=22) (log rank test, P=0.03; Fig. 1A). Patients with the rs2280883 TT genotype (n=216) showed better graft survival than those with the CT or CC genotype (n=15) (P=0.02; Fig. 1B). Patients with the rs3761548 CC genotype (n=209) showed a lower rate of recurrence of the original glomerular disease than those with the AC or AA genotype (n=22) (P=0.01; Fig. 2).
In our study, both the rs3761548 AC and AA genotypes were associated with inferior graft survival and recurrence of primary renal disorders. The Foxp3 rs3761548 AA genotype has been associated with psoriasis [21], unexplained recurrent spontaneous abortion in Chinese individuals [22], and intractability of Graves' disease in Japanese individuals [23]. Recently, an association between the AA genotype and allograft rejection has been reported in kidney transplantation in Chinese individuals [16]. In addition, an association between the rs3761548 A allele and AR and between the rs3761548 AA genotype and lower graft survival have been reported in Indian individuals [18], which is similar to our findings. Polymorphisms in the promoter of the Foxp3 may alter the binding specificity of transcription factors, thus influencing transcription initiation, and therefore, might affect the function or quantity of Treg [24]. Oda et al [7] suggested that the rs3761548 AA genotype leads to a loss of binding with E47 and c-Myb, resulting in defective transcription of Foxp3. Qiu et al [16] demonstrated that patients with the AA genotype are more prone to allograft rejection in kidney transplantation and that Treg function is weaker in patients with the AA genotype than in those with the CC genotype.
The association of Foxp3 SNPs with recurrence of primary renal disorder has yet to be studied. The association of the Th1 response (high production of tumor necrosis factor-α and low production of interleukin-10) with a lower recurrence of IgA nephropathy following kidney transplantation has been reported [25]. The Treg response defect due to Foxp3 SNPs may affect the recurrence of primary glomerular disease. Further studies are needed to elucidate the association between various T cell responses and the recurrence of primary glomerular disease.
In our study, both the rs2280883 CT and CC genotypes were associated with inferior graft survival. The incidence of the Foxp3 rs2280883 CC genotype has been shown to be increased in infertile women without endometriosis in Brazil [26] and Graves's disease in China [27]; however, it was not associated with Graves' disease and juvenile idiopathic arthritis in the United Kingdom [28, 29] or with psoriasis in China [21]. To date, no study has analyzed the association of rs2280883 polymorphisms with allograft outcome following kidney transplantation, although polymorphisms of other genes relevant to host immune responses, such as FasL or IL-17, have been reported [30, 31]. Further studies involving a larger number of patients and other ethnic groups are needed to confirm the association of the rs2280883 CC genotype with renal allograft outcome.
In conclusion, our study revealed the association between the rs3761548 CC and rs2280883 TT genotypes and superior graft survival in kidney transplantation. These findings may help elucidate the role of Treg in kidney transplantation and predict renal allograft outcome.
Acknowledgments
This study was supported by grant number 0420140450 (2014-1187) from the SNUH research fund.
References
1. Yates PJ, Nicholson ML. The aetiology and pathogenesis of chronic allograft nephropathy. Transpl Immunol. 2006; 16:148–157. PMID: 17138047.
2. Zheng XX, Sánchez-Fueyo A, Sho M, Domenig C, Sayegh MH, Strom TB. Favorably tipping the balance between cytopathic and regulatory T cells to create transplantation tolerance. Immunity. 2003; 19:503–514. PMID: 14563315.
3. Li XC, Turka LA. An update on regulatory T cells in transplant tolerance and rejection. Nat Rev Nephrol. 2010; 6:577–583. PMID: 20683480.
4. Graca L, Cobbold SP, Waldmann H. Identification of regulatory T cells in tolerated allografts. J Exp Med. 2002; 195:1641–1646. PMID: 12070291.
5. Nagahama K, Nishimura E, Sakaguchi S. Induction of tolerance by adoptive transfer of Treg cells. Methods Mol Biol. 2007; 380:431–442. PMID: 17876110.
6. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003; 4:330–336. PMID: 12612578.
7. Oda JM, Hirata BK, Guembarovski RL, Watanabe MA. Genetic polymorphism in FOXP3 gene: imbalance in regulatory T-cell role and development of human diseases. J Genet. 2013; 92:163–171. PMID: 23640423.
8. He Y, Na H, Li Y, Qiu Z, Li W. Foxp3 rs3761548 polymorphism predicts autoimmune disease susceptibility: a meta-analysis. Hum Immunol. 2013; 74:1665–1671. PMID: 23993983.
9. Bestard O, Cruzado JM, Rama I, Torras J, Gomà M, Serón D, et al. Presence of FoxP3+ regulatory T cells predicts outcome of subclinical rejection of renal allografts. J Am Soc Nephrol. 2008; 19:2020–2026. PMID: 18495961.
10. Grimbert P, Mansour H, Desvaux D, Roudot-Thoraval F, Audard V, Dahan K, et al. The regulatory/cytotoxic graft-infiltrating T cells differentiate renal allograft borderline change from acute rejection. Transplantation. 2007; 83:341–346. PMID: 17297410.
11. Mansour H, Homs S, Desvaux D, Badoual C, Dahan K, Matignon M, et al. Intragraft levels of Foxp3 mRNA predict progression in renal transplants with borderline change. J Am Soc Nephrol. 2008; 19:2277–2281. PMID: 18667728.
12. Krepsova E, Tycova I, Sekekova A, Wohlfahrt P, Hruba P, Striz I, et al. Effect of induction therapy on the expression of molecular markers associated with rejection and tolerance. BMC Nephrol. 2015; 16:146. PMID: 26286066.
13. Iwase H, Kobayashi T, Kodera Y, Miwa Y, Kuzuya T, Iwasaki K, et al. Clinical significance of regulatory T-cell-related gene expression in peripheral blood after renal transplantation. Transplantation. 2011; 91:191–198. PMID: 21157405.
14. Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005; 353:2342–2351. PMID: 16319383.
15. Veronese F, Rotman S, Smith RN, Pelle TD, Farrell ML, Kawai T, et al. Pathological and clinical correlates of FOXP3+ cells in renal allografts during acute rejection. Am J Transplant. 2007; 7:914–922. PMID: 17286616.
16. Qiu XY, Jiao Z, Zhang M, Chen JP, Shi XJ, Zhong MK. Genetic association of FOXP3 gene polymorphisms with allograft rejection in renal transplant patients. Nephrology (Carlton). 2012; 17:423–430. PMID: 22239151.
17. Engela AU, Boer K, Roodnat JI, Peeters AM, Eilers PH, Kal-van Gestel JA, et al. Genetic variants of FOXP3 influence graft survival in kidney transplant patients. Hum Immunol. 2013; 74:751–757. PMID: 23459079.
18. Misra MK, Mishra A, Pandey SK, Kapoor R, Sharma RK, Agrawal S. Association of functional genetic variants of transcription factor Forkhead Box P3 and Nuclear Factor-κB with end-stage renal disease and renal allograft outcome. Gene. 2016; 581:57–65. PMID: 26794449.
19. Song EY, Park MH, Kang SJ, Park HJ, Kim BC, Tokunaga K, et al. HLA class II allele and haplotype frequencies in Koreans based on 107 families. Tissue Antigens. 2002; 59:475–486. PMID: 12445317.
20. Chen X, Gan T, Liao Z, Chen S, Xiao J. Foxp3 (-/ATT) polymorphism contributes to the susceptibility of preeclampsia. PLoS One. 2013; 8:e59696. PMID: 23560055.
21. Gao L, Li K, Li F, Li H, Liu L, Wang L, et al. Polymorphisms in the FOXP3 gene in Han Chinese psoriasis patients. J Dermatol Sci. 2010; 57:51–56. PMID: 19880293.
22. Wu Z, You Z, Zhang C, Li Z, Su X, Zhang X, et al. Association between functional polymorphisms of Foxp3 gene and the occurrence of unexplained recurrent spontaneous abortion in a Chinese Han population. Clin Dev Immunol. 2012; 2012:896458. PMID: 21876709.
23. Inoue N, Watanabe M, Morita M, Tomizawa R, Akamizu T, Tatsumi K, et al. Association of functional polymorphisms related to the transcriptional level of FOXP3 with prognosis of autoimmune thyroid diseases. Clin Exp Immunol. 2010; 162:402–406. PMID: 20942809.
24. Hoogendoorn B, Coleman SL, Guy CA, Smith K, Bowen T, Buckland PR, et al. Functional analysis of human promoter polymorphisms. Hum Mol Genet. 2003; 12:2249–2254. PMID: 12915441.
25. Coppo R, Amore A, Chiesa M, Lombardo F, Cirina P, Andrulli S, et al. Serological and genetic factors in early recurrence of IgA nephropathy after renal transplantation. Clin Transplant. 2007; 21:728–737. PMID: 17988266.
26. André GM, Barbosa CP, Teles JS, Vilarino FL, Christofolini DM, Bianco B. Analysis of FOXP3 polymorphisms in infertile women with and without endometriosis. Fertil Steril. 2011; 95:2223–2227. PMID: 21481380.
27. Zheng L, Wang X, Xu L, Wang N, Cai P, Liang T, et al. Foxp3 gene polymorphisms and haplotypes associate with susceptibility of Graves' disease in Chinese Han population. Int Immunopharmacol. 2015; 25:425–431. PMID: 25708657.
28. Owen CJ, Eden JA, Jennings CE, Wilson V, Cheetham TD, Pearce SH. Genetic association studies of the FOXP3 gene in Graves' disease and autoimmune Addison's disease in the United Kingdom population. J Mol Endocrinol. 2006; 37:97–104. PMID: 16901927.
29. Eastell T, Hinks A, Thomson W. BSPAR Study Group. SNPs in the FOXP3 gene region show no association with Juvenile Idiopathic Arthritis in a UK Caucasian population. Rheumatology (Oxford). 2007; 46:1263–1265. PMID: 17526924.
30. Fadel FI, Elshamaa MF, Salah A, Nabhan M, Rasheed M, Kamel S, et al. Fas/Fas Ligand pathways gene polymorphisms in pediatric renal allograft rejection. Transpl Immunol. 2016; 37:28–34. PMID: 27109035.
31. Park H, Shin S, Park MH, Kim YS, Ahn C, Ha J, et al. Association of IL-17F gene polymorphisms with renal transplantation outcome. Transplant Proc. 2014; 46:121–123. PMID: 24507036.
Fig. 1
Kaplan-Meier survival analysis of graft survival according to Foxp3 polymorphisms. (A) Patients with rs3761548 CC genotype (n=209) showed better graft survival than those with AC or AA genotype (n=22) (log rank test, P=0.03). (B) Patients with rs2280883 TT genotype (n=216) showed better graft survival than those with CT or CC genotype (n=15) (P=0.02).
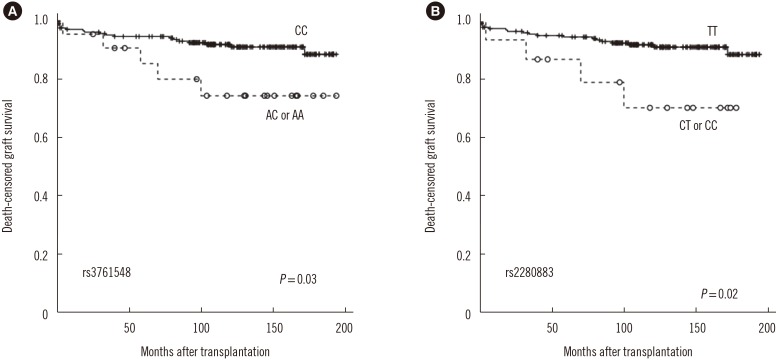
Fig. 2
Kaplan-Meier survival analysis of recurrence of original glomerular disease post kidney transplantation according to Foxp3 polymorphism rs3761548.
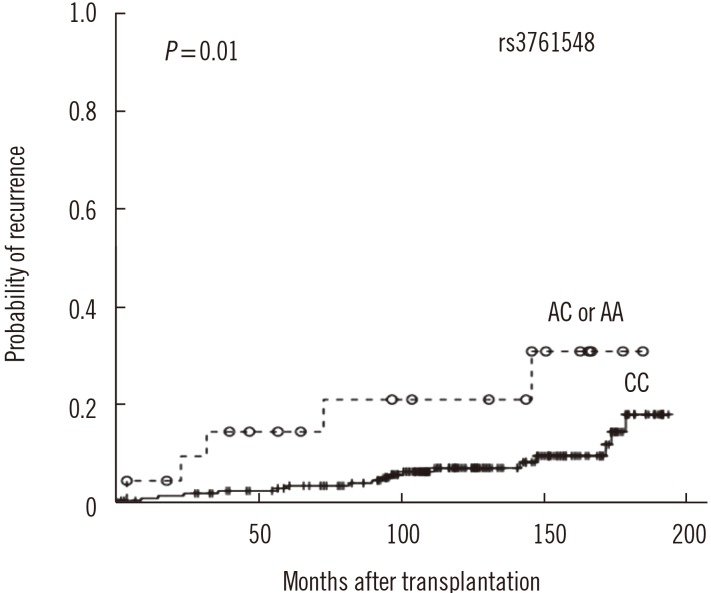
Table 1
Characteristics of the study population and univariate Cox proportional hazards regression analysis with regard to graft survival
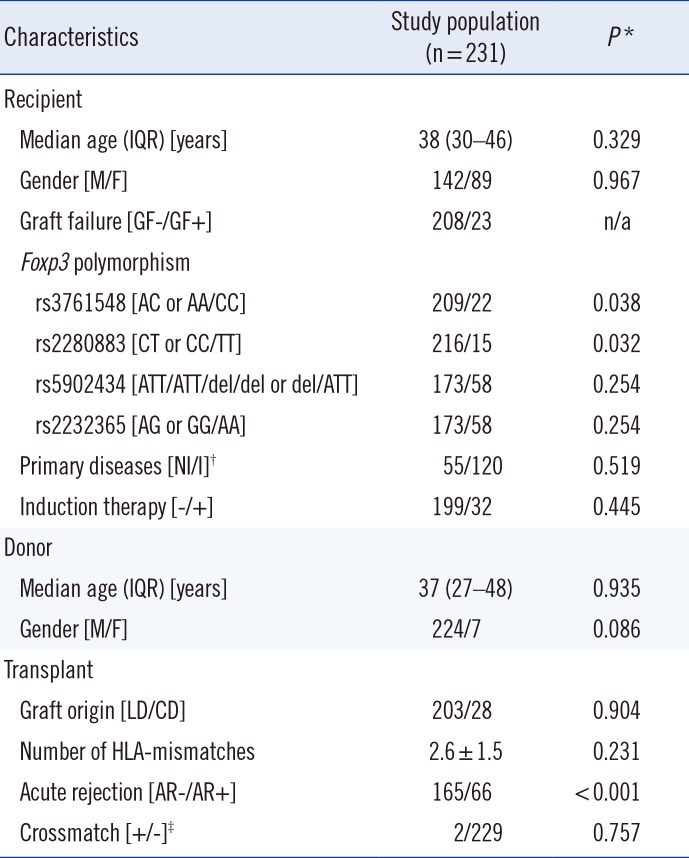
*Univariate Cox regression analysis; †56 (24.2%) cases could not be defined as either primary disease category; ‡All 231 cases were negative for cytotoxic crossmatch, and two cases were positive only for T-cell flowcytometric crossmatch.
Abbreviations: IQR, interquartile range; M, male; F, female; NI, non-inflammatory; I, inflammatory; n/a, not available; LD, living donor; CD, cadaveric donor.
Table 2
Sequence-specific primers for Foxp3 polymorphisms
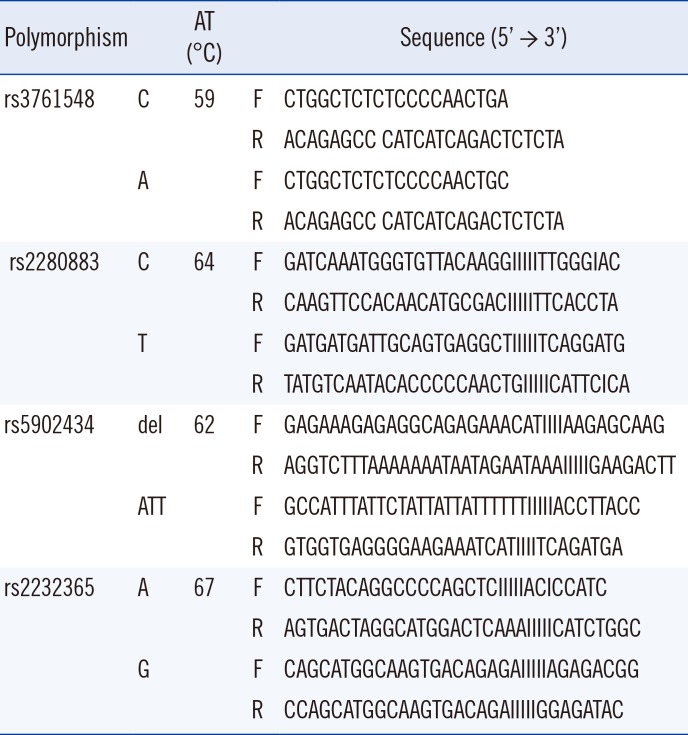
Table 3
Association of Foxp3 polymorphisms with graft rejection




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download