INTRODUCTION
Respiratory viral (RV) infection is one of the most common infectious diseases worldwide [
123]. The rapid and accurate diagnosis of the underlying pathogen is crucial for establishing good clinical practices aimed at reducing morbidity and mortality [
456]. Diagnosing RV infections involves detection of the causative viruses in specimens obtained from patients, such as sputum and/or nasopharyngeal swabs. Traditionally, virus cultures and/or direct fluorescent-antibody assays have been used to detect the viruses [
789]. Recently, multiplex PCR-based methods have been developed and introduced for clinical use, given their advantages of decreased hands-on time and improved sensitivity [
89].
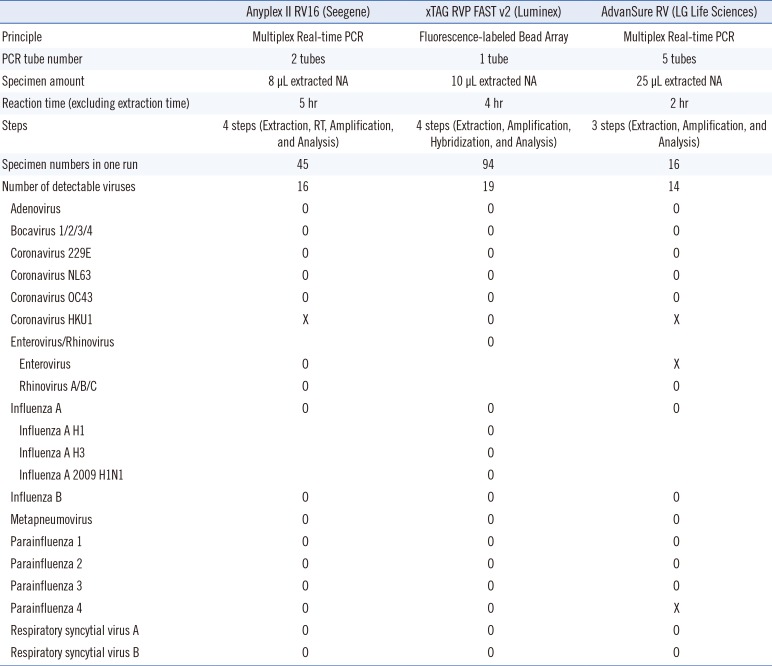
Luminex technology has been widely adopted in clinical laboratories for various tests such as multiplex PCR, human leukocyte antigen tests, and protein assays [
1011]. The Luminex xTAG respiratory viral panel (RVP) and RVP FAST v1 assays (Luminex Molecular Diagnostics, Toronto, Canada) proved to be clinically useful in previous studies [
12131415], and a revised version (xTAG RVP Fast v2) (Luminex Molecular Diagnostics) was recently released in 2013. The Luminex xTAG RVP Fast v2 assay is a multiplex reverse transcriptase (RT)-PCR assay that is incorporated with the Luminex tagging system. Following the first release of the Luminex xTAG RVP assay, Luminex Molecular Diagnostics issued a revised version. The new assay have several advantages over the other assays: 1) detect more types of viruses (19 strains); 2) distinguish among influenza A subtypes (H1, H3, 2009 H1N1), coronavirus HKU1, and parainfluenza type 4; 3) handle the largest number of specimens in one run (94 specimens); 4) use a small specimen volume (10 µL of nucleic acids); and 5) produce semi-quantitative results as the fluorescence intensity.
However, it is unclear whether the new version of the Luminex system has similar or superior virus detection abilities compared with other available multiplex RT-PCR assays.
In the present study, we evaluated the clinical performance of the Luminex xTAG RVP Fast v2 assay for detecting respiratory viruses compared with that of Anyplex II RV16 detection kit (Seegene, Seoul, Korea) and AdvanSure RV real-time RT-PCR assay (LG Life Sciences, Seoul, Korea).
Go to :

DISCUSSION
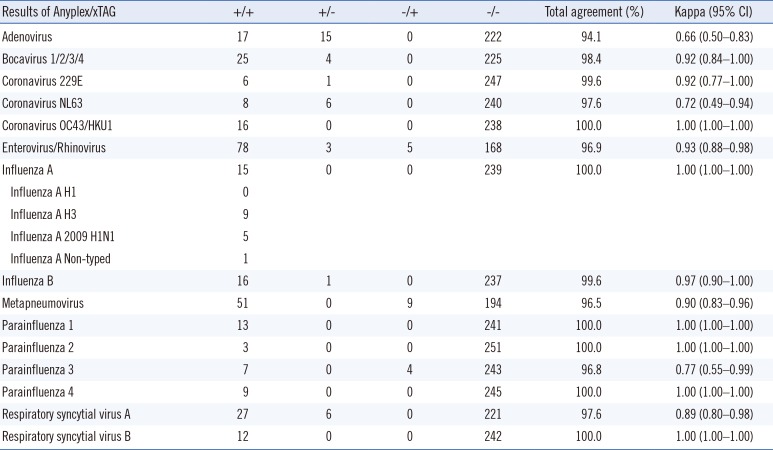
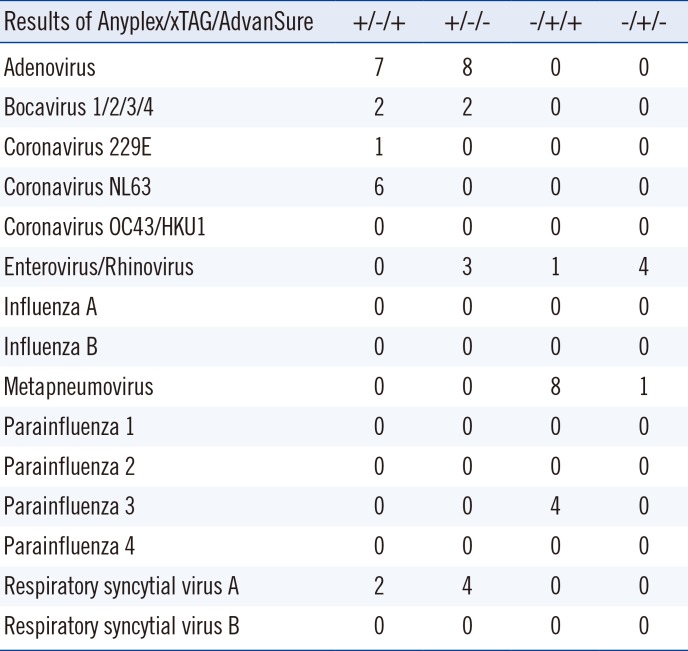
In the present study, we evaluated the ability of the Luminex xTAG RVP Fast v2 assay to detect common respiratory pathogens. Its performance was compared with that of the Anyplex II RV16 assay, and any discrepancies in the results were confirmed by using a third multiplex PCR assay, AdvanSure RV. In general, the Anyplex II RV16 assay detected more pathogens than did the Luminex xTAG RVP Fast v2 assay, which was not surprising considering that only specimens that showed at least one positive result by the Anyplex II RV16 assay were collected for this comparison. Nonetheless, the Luminex xTAG RVP v2 assay detected more metapneumovirus and parainfluenza type 3 specimens.
The xTAG RVP v2 and Anyplex II RV16 assays showed relatively poor concordance for adenovirus, coronavirus NL63, and parainfluenza type 3 specimens. Taking into account the results of the third assay, we found that the Anyplex II RV16 assay detected more adenovirus, bocavirus, and respiratory syncytial virus type A specimens. Moreover, metapneumovirus, parainfluenza type 3, enterovirus, and rhinovirus were more likely to be detected by the Luminex xTAG RVP Fast v2 assay.
The virus culture supernatants were also tested to evaluate the abilities of the kits to detect various strains. In these experiments, none of the three kits was able to detect all of the adenovirus strains. As there are more than 60 serotypes of adenoviruses [
1718], it may be challenging to design assays that can detect all of the subtypes. In addition, the AdvanSure RV assay classified the enterovirus as a rhinovirus because this assay was not originally designed to detect enteroviruses.
Among the 254 specimens tested, 92 showed multiple infections with either the Anyplex II RV16 assay or the Luminex xTAG RVP Fast v2 assay. For cases with multiple infections, discrepancies between the two assays were revealed in 40 cases (43.5%). The discrepancy mainly resulted from the inability to detect accompanying viruses, mostly adenovirus (13 cases), metapneumovirus (7 cases), and coronavirus NL 63 (6 cases). Among the specimens with three complete sets of assay results, there were slightly more discrepancies for cases of multiple infections than for single-infection cases (26 vs 17). However, we were unable to examine this difference statistically owing to the small sample size.
The discrepant results might be due to several factors, including differences in the target genes and primers and differences in the analytical sensitivities of the assays. Previous studies have shown that different targets for amplification can affect the analytical sensitivities and specificities of assays [
1219].
In contrast to the Anyplex II RV16 assay, the Luminex xTAG RVP FAST v2 assay could differentiate among three subtypes of the influenza A virus; thus, this assay may be more helpful, especially during the influenza season. Moreover, only the Luminex xTAG RVP Fast v2 assay could distinguish between coronavirus OC43 and HKU1 specimens. However, it should be noted that it was challenging to directly compare the detection capabilities of the assays because Luminex xTAG RVP Fast v2 reports its capabilities in units of copies/mL, whereas the other assays report the results in units of tissue culture infective dose 50 (TCID50)/mL. Therefore, laboratory personnel should be aware of the characteristics and differences of the assays they use and should be cautious when performing the assays and reporting the results. In particular, laboratory staff and physicians should maintain close communication to ensure the best use of the assays.
For the specimens with discordant results, many showed weak positive reactions (“+” reactions, based on the cycles at which the melting curve analyses revealed the presence of the targets) in the initial Anyplex II RV16 assay. A previous study using serial dilutions of plasmid standards of several viruses showed that greater discrepancies existed in low-copy specimens [
13]. However, the cycle threshold values for the AdvanSure RV assay and the fluorescence intensities for the Luminex xTAG RVP Fast v2 assays were highly variable; thus, the discrepant results may be related, at least in part, to the weak positive reactions, although other unexplained factors may also be involved. Future studies should investigate these possibilities.
Several studies have investigated the clinical utility of the Luminex xTAG RVP assay for detection of respiratory pathogens. While the Luminex assay showed generally higher sensitivities than traditional assays, it could detect more influenza, respiratory syncytial virus, metapneumovirus, and rhinovirus/enterovirus cases, while detect less adenovirus and coronavirus cases [
12131415]. And some have reported that great discrepancies were discovered for adenovirus and influenza B virus cases.
Similar results were obtained in the present study, with the Luminex xTAG RVP assay displaying high detection capabilities for detecting rhinovirus/enterovirus and metapneumovirus cases but low detection capabilities for coronavirus cases. In fact, many commercial kits tend to produce discrepant results for adenovirus specimens [
2021].
The main limitation of this study is that we chose our specimens retrospectively; thus, selection bias may have existed and affected the results. In addition, we were unable to assure the stability of the stored specimens. Indeed, some of the specimens showed discrepant results between the initial and follow-up Anyplex II RV16 assays. As sequencing analysis was not performed for the specimens, we compared the results of the assays for concordance, making it difficult to directly determine the analytical sensitivity and/or specificity of these assays.
In conclusion, we evaluated the clinical utility of the Luminex xTAG RVP Fast v2 assay. We found that this assay could detect various respiratory pathogens simultaneously and that the performance was comparable to that of the existing methods in terms of agreement. The Luminex xTAG RVP Fast v2 assay may be a good alternative method in clinical laboratories for the diagnosis of RV infections.
Go to :






 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download