Abstract
Background
We evaluated the performance of the BD MAX StaphSR Assay (SR assay; BD, USA) for direct detection of Staphylococcus aureus and methicillin resistance not only in S. aureus but also in coagulase-negative Staphylococci (CNS) from positive blood cultures.
Methods
From 228 blood culture bottles, 103 S. aureus [45 methicillin-resistant S. aureus (MRSA), 55 methicillin-susceptible S. aureus (MSSA), 3 mixed infections (1 MRSA+Enterococcus faecalis, 1 MSSA+MRCNS, 1 MSSA+MSCNS)], and 125 CNS (102 MRCNS, 23 MSCNS) were identified by Vitek 2. For further analysis, we obtained the cycle threshold (Ct) values from the BD MAX system software to determine an appropriate cutoff value. For discrepancy analysis, conventional mecA/mecC PCR and oxacillin minimum inhibitory concentrations (MICs) were determined.
Results
Compared to Vitek 2, the SR assay identified all 103 S. aureus isolates correctly but failed to detect methicillin resistance in three MRSA isolates. All 55 MSSA isolates were correctly identified by the SR assay. In the concordant cases, the highest Ct values for nuc, mecA, and mec right-extremity junction (MREJ) were 25.6, 22, and 22.2, respectively. Therefore, we selected Ct values from 0-27 as a range of positivity, and applying this cutoff, the sensitivity/specificity of the SR assay were 100%/100% for detecting S. aureus, and 97.9%/98.1% and 99.0%/95.8% for detecting methicillin resistance in S. aureus and CNS, respectively.
The observation of gram-positive cocci in clusters from positive blood cultures provides only a presumptive identification of Staphylococcus spp. Confirmatory identification and susceptibility testing of Staphylococci recovered from blood cultures require about 48 hr, delaying the administration of pathogen-directed therapy, promoting the unnecessary use of vancomycin, and increasing the pressure to select resistant organisms [1]. Although coagulase-negative Staphylococci (CNS) are generally considered normal skin flora, these microbes can also cause bacteremia in critically ill patients [2], especially causing catheter-related infections [3]. Thus, rapid and accurate detection of methicillin resistance is also needed.
The BD Max Staph SR assay (BD Diagnostics, Sparks, MD, USA) is a fully automated, qualitative, in vitro diagnostic test for the direct detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) from nasal swabs. The kit targets mec right-extremity junction (MREJ) (containing the right-extremity of SCCmec and orfX, chromosomal S. aureus gene), the nuc gene encoding a thermostable S. aureus nuclease, genes for methicillin resistance mecA/mecC, and includes a sample-processing control. MREJ detection distinguishes MRSA infection from mixed infections of methicillin-susceptible S. aureus (MSSA) and MRCNS. For instance, to be MRSA-positive, signals for both mecA/mecC and MREJ should be positive, and the nuc gene target may or may not be detected since in rare instances, the nuc gene may be absent in MRSA [4]. A positive result for mecA and nuc genes accompanied by a negative MREJ result indicates a mixed infection of MSSA and MRCNS. The kit detects 11 of 21 types of MREJ (type i, ii, iii, iv, v, vi, vii, ix, xiii, xiv, and xxi), which represents most mecA- and mecC-harboring MRSA strains [1, 5]. A positive result of S. aureus and mecA/mecC indicates MRSA. The assay uses TaqMan probes labeled with fluorescent dyes and the amount of fluorescence detected in the optical channels is directly proportional to the quantity of the corresponding probe that is hydrolyzed. It indicates S. aureus and MRSA positivity if a fluorescent signal is detected for both MREJ and mecA or mecC. The assay indicates S. aureus positivity and MRSA negativity when a fluorescent signal is detected for the nuc gene target only, or if signal is detected for nuc and mecA/mecC in the absence of MREJ (indicative of a S. aureus strain co-colonized with MRCNS). This assay is Conformité Européene (CE) marked and U.S. Food and Drug Administration (FDA)-cleared for the direct detection and differentiation of S. aureus and MRSA from nasal swabs.
In this study, we analyzed the performance of the BD MAX StaphSR Assay (SR assay) for detecting S. aureus and methicillin resistance not only in S. aureus but also in CNS from positive blood culture broths.
A total of 228 consecutive positive blood culture broths (aerobic/anaerobic bottles), one per patient, showing growth of gram-positive cocci in clusters were collected from two hospitals in Korea (Seoul St. Mary's Hospital and Uijeongbu St. Mary's Hospital) from January to October 2015. Bacterial species were identified with a Vitek 2 system and/or Vitek-MS system (bioMérieux, Lyon, France). A 0.5-mL aliquot of positive blood culture broth was taken after the bottle was flagged positive, stored for up to 72 hr at 4℃ prior to testing, and 10 µL was used for the BD Max SR assay. The remaining broths were frozen at -70℃ for further analysis. The number of collected blood culture broths was controlled to represent S. aureus and CNS in approximately equal numbers.
For conventional susceptibility tests, specimens were subcultured on 5% sheep's blood agar plates in a 35℃ incubator for 18-24 hr, and antimicrobial susceptibility tests were performed with the Vitek 2 system. For discrepant cases, latex agglutination for PBP2a and oxacillin minimum inhibitory concentrations (MICs) were determined by the agar dilution method according to the CLSI guideline (M100-S25) [6], and conventional PCR for mecA/mecC was performed according to the protocol recommended by the European Union Reference Laboratory; antimicrobial resistance [7]. After discrepancy analysis, the sensitivity and specificity of the SR assay was calculated. The 95% confidence interval (CI) of the rate was calculated with the Clinical Calculator 1 (http://vassarstats.net/clin1.html). This study was based on the analysis of remaining samples from clinical tests, and the institutional review board approved this study.
Among the 228 blood culture bottles tested, 103 S. aureus [45 MRSA, 55 MSSA, 3 cases of mixed infections (1 MRSA+Enterococcus faecalis, 1 MSSA+MRCNS, 1 MSSA+MSCNS)], and 125 CNS (102 MRCNS, 23 MSCNS) were identified by Vitek 2. The SR assay identified all 103 S. aureus isolates and their methicillin susceptibility correctly. None of the 125 CNS cases were misidentified as S. aureus. Of the 45 MRSA cases identified by Vitek 2, 42 cases were identified as MRSA but three were misidentified as MSSA by the SR assay. All 55 MSSA were identified by Vitek 2 and the SR assay. Therefore, compared with the Vitek 2 results, the agreement rate for S. aureus identification was 100% and that for MRSA and MSSA were 93.5% (43/46, which includes a mixed infection of MRSA) and 100% (57/57), respectively. During the evaluation period, invalid results due to PCR inhibition were not observed in any case.
For further analysis, we obtained Ct values from the BD MAX system software. Ct values range from -1 (meaning negative) to various positive Ct values. The Ct values for the nuc target were <27 for all 103 S. aureus-positive broths (range; 15.6-25.6); for the 125 CNS cases, the Ct value was -1 for 123 cases; and for the remaining two cases, it was 34.1 and 35.9 (Fig. 1).
For the 45 cases of MRSA-only identified by Vitek 2, the Ct value of mecA ranged from 1 to 22.2 for all the 45 cases; the range of Ct values of MREJ was 16.1-22 for the 42 cases; and -1 in three MRSA cases (Fig. 2).
For the three isolates showing MREJ Ct values of -1, the oxacillin MICs were ≥64 µg/mL for all the three isolates and they were positive for mecA. Therefore, the false negative results in three isolates were due to false negative MREJ signals. We sent these samples to BD for MREJ typing, but these isolates were untypable. Taking together the Ct values from the concordant cases, the highest Ct values for nuc, mecA, and MREJ were 25.6, 22, and 22.2, respectively.
For the 55 MSSA cases identified by Vitek 2, the Ct values of mecA were -1 for 44 cases and 24.9-38 for 11 cases. The Ct values of MREJ were -1 for 45 cases, 27.8-36.5 for nine cases, and one isolate showed a Ct value of 17.6. (Fig. 2). However, all cases yielded a result of -1 for either mecA or MREJ, indicating a negative result for methicillin resistance by the SR assay. The results of mecA PCR and oxacillin MIC of the 11 cases are summarized in Table 1. Two isolates (strains 43 and 119) were mecA-positive, oxacillin susceptible (their mecA Ct values by the SR assay were 26.8 and 32.3, respectively), and one isolate (strain no. 120) was mecA-positive oxacillin-heteroresistant (Ct value for mecA and MREJ was 26.6 and -1, respectively). MREJ typing of this heteroresistant isolate was MREJ type XX, which is not included in the SR assay. We presumed that mecA results from the remaining eight cases were false positive due to non-specific amplification, and the MSSA case showing an MREJ Ct value of 17.6 might be an empty cassette strain.
MREJ Ct values were -1 in all 102 MRCNS cases, and the mecA Ct values ranged from 16.8 to 24 for 100 cases, leaving two cases with values of -1 and 30.1 (Fig. 1). For the isolate with a mecA Ct value of -1, mecA PCR was negative and the oxacillin MIC was 0.25 µg/mL. Therefore, it was MSCNS and misidentified as MRCNS by Vitek 2. For the isolate with a mecA Ct value of 30.1, mecA PCR was positive and oxacillin MIC was >64 µg/mL.
For 23 MSCNS cases, all isolates showed a MREJ Ct value of -1, but for the mecA gene, 16 isolates showed Ct values of -1 and the remaining seven isolates showed mecA Ct values of 21.3, 31.1, 34.6, 35, 37.1, 37.8, and 39.2 (Fig. 1). Of these seven isolates, one isolate (No. 13, mecA Ct of 21.3) was mecA-positive but susceptible to oxacillin (MIC 0.25 µg/mL).
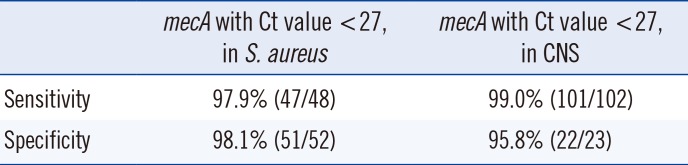
Compared with that of the conventional mecA PCR, with a Ct cutoff <27, sensitivity of the SR assay for methicillin resistance (mecA) was 98.7% [147/149, 45 MRSA+2 mecA positive MSSA+1 heteroresistant MRSA+101 MRCNS]. The specificity for detecting methicillin resistance was 97.4% (74/76, 52 MSSA+24 MSCNS). For S. aureus, the sensitivity for detecting mecA was 97.9% (47/48, 95% CI 87.5-99.9%, 45 MRSA+2 mecA positive MSSA+1 mecA positive heteroresistant MRSA), and the specificity was 98.1% (51/52, 95% CI 88.4-99.9%). For CNS, sensitivity/specificity for detecting mecA was 99.0% (100/101, 95% CI 93.8-99.9%)/95.8% (23/24, 95% CI 76.9-99.8%). Without applying the new cutoff value suggested by our study, the sensitivity and specificity of the SR assay for detecting mecA-positive S. aureus were 87.5% (42/48) and 100% (52/52), respectively (Table 2).
The BD MAX SR assay was excellent for detecting S. aureus irrespective of the presence of a mixed infection. Currently, the Ct values of the tests are not shown in the result report of the SR assay. In this study, we obtained Ct values from BD and found that the Ct value distribution for the nuc gene was clearly separate, and the highest Ct value among the S. aureus was 25.6 including mixed infections. This is in line with another study where the Ct values for the nuc target were <27 for all MRSA-positive blood cultures [8]; therefore, we selected a Ct value <27 to indicate positivity.
The SR assay makes use of the combined interpretation of two targets [SCCmec-orfX (MREJ) and mecA/mecC] to differentiate MRSA colonization and mixed colonization of MSSA and MRCNS in nasal swab specimens. However, from the positive blood culture broths, mixed cultures of S. aureus and CNS were observed only in two cases (0.9%) and despite the high oxacillin resistance rate (about 60%) among CNS in our hospital, a mixed case of MSSA and MRCNS was observed only once (0.4%). MREJ was not detected in 6.7% (3/45) of MRSA isolates in this study, which is in line with a previous study from the U.S. where 8.3% (3 of 36) of MRSA from positive blood cultures were strains with MREJ types that could not be detected by the BD GeneOhm Staph SR assay [1]. In an Australian study, the sensitivity of the BD GeneOhm Staph SR for detecting MRSA was 50%, based on the prevailing SCCmec types, and they recommend the nuc/mec assay, because MREJ detection will always be vulnerable to epidemiological variations [9]. In addition, in the nine MSSA cases, MREJ showed a positive signal although the Ct values were >27. Our results agree with those reported by Thomas et al [9], and we presume that the use of two targets (nuc and mecA) excluding MREJ will increase the sensitivity in detecting MRSA in positive blood cultures without decreasing specificity.
To our knowledge, this is the first study to investigate the usefulness of the SR assay in detecting methicillin resistance in CNS from blood culture broths. With the Ct value positivity range of 0-27, the sensitivity and specificity for detecting oxacillin resistance was very high for both S. aureus and CNS. The newly suggested cutoff value provided enhanced sensitivity compared with that by the results of the current SR assay. Considering the fact that CNS also causes bacteremia in immune-compromised patients, this could be very useful for managing them.
We also found that 2% (2/100) of S. aureus and 0.8% (1/125) of CNS cases were mecA-positive and oxacillin-susceptible. The mecA Ct values of these isolates were 24.9, 26.8, and 21.3. In Taiwan, among S. aureus with oxacillin MICs of 2 and 1 µg/mL, as determined by the Sensititre broth microdilution test, 57.1% and 3.3% were mecA-positive, respectively, and belonged to a dominant Taiwan C-MRSA clone (clonal complex [CC]59); most of them were the SCCmec type V [10]. In a recent study from the U.S., the prevalence of mecA-positive, oxacillin-susceptible S. aureus was approximately 3% and the rate of reversion to resistance was at a modest frequency (approximately 10-7), which is the same order of magnitude as the frequency of spontaneous resistance to drugs like rifampicin [11]. Although combination therapy using a β-lactam and a second antibiotic suppressing the small revertant population may be superior to vancomycin, mecA-positive MSSA can revert to MRSA [11] and current guidelines recommend that these strains be reported as resistant. Further studies are needed for the appropriate treatment of infections caused by these strains.
In conclusion, with the Ct range of 0-27 for positivity by the nuc/mec assay, the SR assay is useful for the rapid detection of not only MRSA but also MRCNS in blood cultures. We suggest not using the MREJ target because it is vulnerable to epidemiologic variation and co-infection with CNS is rare in positive blood cultures. In addition, for the accurate detection of mecA-positive, oxacillin-susceptible isolates, genotypic detection of mecA is needed.
Acknowledgments
BD (BD Diagnostics, New Jersey, USA) provided BD Max assay kits for this study but was not involved in either data collection or preparation of the manuscript.
References
1. Snyder JW, Munier GK, Heckman SA, Camp P, Overman TL. Failure of the BD GeneOhm StaphSR assay for direct detection of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates in positive blood cultures collected in the United States. J Clin Microbiol. 2009; 47:3747–3748. PMID: 19794051.
2. Savithri MB, Iyer V, Jones M, Yarwood T, Looke D, Kruger PS, et al. Epidemiology and significance of coagulase-negative staphylococci isolated in blood cultures from critically ill adult patients. Crit Care Resusc. 2011; 13:103–107. PMID: 21627578.
3. Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009; 23:73–98. PMID: 19135917.
4. Pasanen T, Korkeila M, Mero S, Tarkka E, Piiparinen H, Vnopio-Varkila J, et al. A selective broth enrichment combined with real-time nuc-mecA-PCR in the exclusion of MRSA. Apmis. 2010; 118:74–80. PMID: 20041874.
BD MAX StaphSR [package insert]. Catalog no. 442999, publication P0153 (01). BD Diagnostics Sparks. MD, USA: 2013.
6. CLSI. Performance standards for antimicrobial susceptibility testing. M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute;2015.
7. EURL-AR. European union reference laboratory antimicrobial resistance. Protocol for PCR amplification of MECA, MECC (MECALGA251), SPA and PVL. Lyngby, Denmark: DTU Food, National Food Institute;2012.
8. Dalpke AH, Hofko M, Hamilton F, Meckenzie L, Zimmermann S, Templeton K. Evaluation of the BD Max StaphSR Assay for rapid identification of Staphylococcus aureus and Methicillin-resistant S. aureus in Positive Blood Culture Broths. J Clin Microbiol. 2015; 53:3630–3632. PMID: 26292311.
9. Thomas L, van Hal S, O'Sullivan M, Kyme P, Iredell J. Failure of the BD GeneOhm StaphS/R assay for identification of Australian methicillin-resistant Staphylococcus aureus strains: duplex assays as the "gold standard" in settings of unknown SCCmec epidemiology. J Clin Microbiol. 2008; 46:4116–4117. PMID: 18945832.
10. Chen FJ, Huang IW, Wang CH, Chen PC, Wang HY, Lai JF, et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012; 50:1679–1683. PMID: 22378906.
11. Proulx MK, Palace SG, Gandra S, Torres B, Weir S, Stiles T, et al. Reversion From Methicillin Susceptibility to Methicillin Resistance in Staphylococcus aureus During Treatment of Bacteremia. J Infect Dis. 2016; 213:1041–1048. PMID: 26503983.
Fig. 1
Distribution of Ct values of nuc gene among the S. aureus and coagulase-negative Staphylococci (A) and mecA/mecC among methicillin-resistant and methicillin-susceptible coagulase-negative Staphylococci (B). The nuc gene assay perfectly distinguishes CNS from SA. Note that most of MRCNS reveal Ct value under 27.
Abbreviations: Ct, cycle threshold; SA, Staphylococcus aureus; CNS, coagulase-negative Staphylococcus; MSCNS, methicillin susceptible CNS; MRCNS, methicillin resistant CNS.
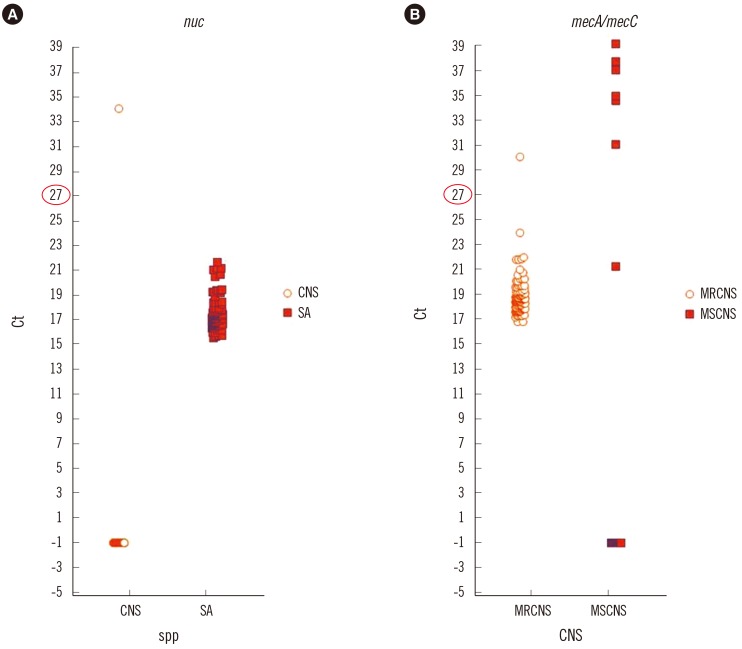
Fig. 2
Distribution of MREJ (A) and mecA/mecC (B) Ct values among methicillin-resistant and methicillin-susceptible S. aureus. Note that there are MREJ negative MRSAs.
Abbreviations: Ct, cycle threshold; SA, Staphylococcus aureus; MSSA, methicillin susceptible SA; MRSA, methicillin resistant SA; MREJ, mec right-extremity junction.
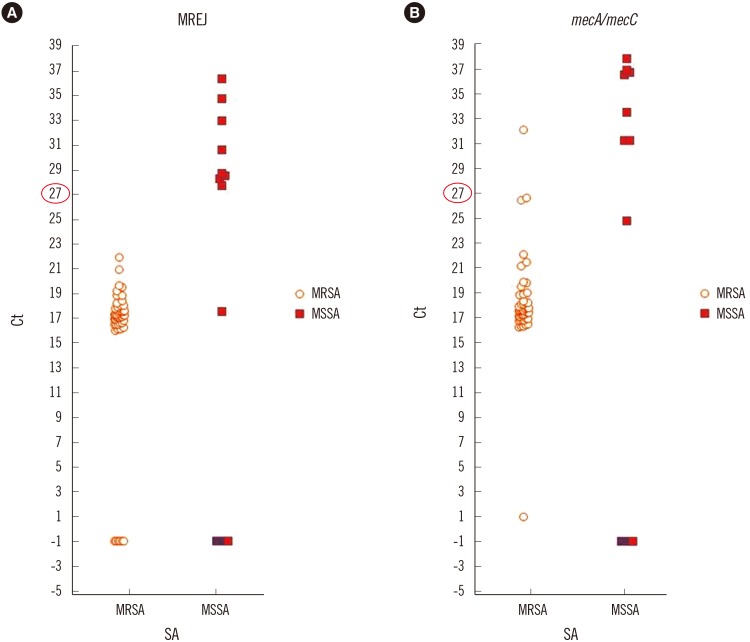
Table 1
Characteristics of strains identified as MSSA by Vitek 2, but with positive mecA/mecC Ct values according to the SR assay
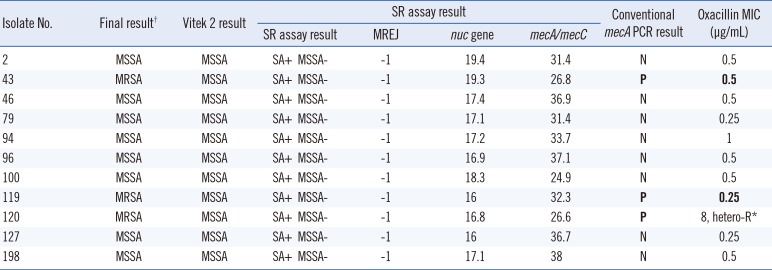
There are mecA-positive and oxacillin-susceptible isolates, but they are considered as MRSA. MIC criteria for MRSA is ≥4 µg/mL with oxacillin.
*hetero-R=heteroresistance; †Final results after resolving discrepancies between Vitek 2 and SR assay with conventional mecA/mecC PCR.
Abbreviations: SA, Staphylococcus aureus; MSSA, methicillin susceptible SA; MRSA, methicillin resistant SA; MREJ, mec right-extremity junction; N, negative; P, positive.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download