Dear Editor,
Robinsoniella peoriensis is a gram-positive, spore-forming, anaerobic rod originally isolated from swine manure storage pits [1]. Most R. peoriensis strains have been isolated from environmental sources, but this organism was isolated from human blood, abdominal fluid, wound, muscle hematoma puncture, or necrotic tissue in nine cases [23456]. Here, we describe a second Korean case of R. peoriensis bacteremia, which was confirmed via 16S rRNA sequencing.
A 63-yr-old man with diabetes mellitus and multiple lacunar infarctions was admitted for scheduled chemotherapy. He had been diagnosed as having small cell lung cancer six months prior to admission. He lived in an urban area of Seoul and denied having traveled or having had any animal contact in the past year. His initial vital signs on admission were normal, and initial laboratory test results were as follows: Hb, 67 g/L; white blood cell count, 6.3×109/L (neutrophils, 78.4%); and platelets, 87×109/L. The serum C-reactive protein (CRP) level was elevated to 108.10 nmol/L, and the lactate dehydrogenase (LDH) level was elevated to 13.83 µkat/L. The patient was suspected of having aspiration pneumonia because of haziness in the right middle and lower lobe fields on chest X-ray. Blood culture sets (two aerobic and one anaerobic) were taken from three separate vein sites. The patient was initially treated with 4 g Tazocin via intravenous administration three times per day.
On day 3 after admission, the patient developed fever of 38.4℃. On day 4 after admission, the body temperature was elevated to 39.5℃, the CRP level was elevated to 253.72 nmol/L, and chest X-ray revealed consolidations with increased patchiness in both lung fields.
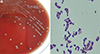
After one day of incubation, one anaerobic blood culture showed growth of microorganisms; two days later, gray-white, smooth, and non-hemolytic colonies of unequal sizes were observed only on the anaerobic blood plate (Fig. 1A). Gram staining of a purified colony indicated the presence of gram-positive, rod-shaped bacteria with ovoid-shaped spores located centrally (Fig. 1B). The BD BACTEC FX (BD Diagnostics, Heidelberg, Germany) and VITEK2 (bioMérieux, Marcy l'Etoile, France) systems indicated the presence of Clostridium clostridioforme (92%), with a questionable level of confidence owing to its low biofrequency. An antimicrobial susceptibility test of the isolate could not be performed. On day 5 after admission, antibiotic therapy with Tazocin was switched to imipenem at 500 mg intravenous (IV) three times per day. No organisms were grown on follow-up blood culture. However, the patient died from progression of pneumonia and respiratory failure. The etiologic diagnosis was established post-mortem by means of molecular analysis. To identify the bacteria, molecular identification was performed by DNA amplification and sequencing analysis of the 16S rRNA gene [8]. A GenBank BLAST search revealed that the 16S rRNA gene sequence of the isolate showed 99.17% homology for 1,316 bp with the corresponding sequence of R. peoriensis (GenBank accession number NR_041882.1). Murimonas intestini, C. oroticum, and Hespellia porcina were the next best matches, with similarities of 94.60%, 94.53%, and 94.47%, respectively. When the sequence was submitted to the Ez-Taxon database v2.1 (http://www.ezbiocloud.net), the highest similarity was with R. peoriensis (99.04%), followed by C. nexile, C. saccharolyticum, and M. intestini, with similarities of 94.88%, 94.33%, and 94.26%, respectively. A phylogenetic tree was constructed by using the neighbor-joining method in Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 (http://www.megasoftware.net; Fig. 2).
Current phenotypic identification systems, including the VITEK2 system, are unable to identify R. peoriensis and may confuse it with other organisms such as Clostridium [4567]. An increasing number of studies have recently emerged describing microorganisms misidentified by conventional methods [9]. Therefore, identification of rare bacteria and bacteria with unusual phenotypic profiles by conventional methods needs to be confirmed by a reliable tool such as 16S rRNA sequencing [10].
We report our experience with the use of 16S rRNA sequencing and various databases for the identification of an unknown anaerobic organism. This report represents the second case of R. peoriensis isolation from human blood in Korea. Most anaerobic infections originate from the patient's own microflora or another exogenous environmental source, and anaerobic bacteremia may be serious or even life-threatening in immunocompromised patients. Although R. peoriensis is rarely reported as a pathogen, the possibility should be carefully evaluated depending on the patient's status. In the present case, the patient suffered from a chronic debilitation disorder with diabetes mellitus and lung cancer while undergoing chemotherapy. R. peoriensis isolated from a blood specimen was successfully identified by using molecular methods in a clinical laboratory.
Figures and Tables
References
1. Cotta MA, Whitehead TR, Falsen E, Moore E, Lawson PA. Robinsoniella peoriensis gen. nov., sp. nov., isolated from a swine-manure storage pit and a human clinical source. Int J Syst Evol Microbiol. 2009; 59:150–155.

2. López P, Belda S, García M, Royo G. Infection of a spontaneous muscular hematoma due to Robinsoniella peoriensis, in a patient with alcoholic liver cirrhosis. Enferm Infecc Microbiol Clin. 2010; 28:565–567.
3. Cassir N, Laget L, Renvoisé A, Gennari JM, Drancourt M. Robinsoniella peoriensis infection following surgery for scoliosis: a case report. J Med Case Rep. 2012; 6:174.

4. Gomez E, Gustafson DR, Colgrove R, Ly T, Santana R, Rosenblatt JE, et al. Isolation of Robinsoniella peoriensis from four human specimens. J Clin Microbiol. 2011; 49:458–460.

5. Shen D, Chen R, Ye L, Luo Y, Tang YW. Robinsoniella peoriensis bacteremia in a patient with pancreatic cancer. J Clin Microbiol. 2010; 48:3448–3450.
6. Jeon Y, Kim TS, Kim HB, Park KU, Song J, Kim EC. First Korean case of Robinsoniella peoriensis bacteremia in a patient with aspiration pneumonia. Ann Lab Med. 2012; 32:370–374.

7. Ferraris L, Aires J, Butel MJ. Isolation of Robinsoniella peoriensis from the feces of premature neonates. Anaerobe. 2012; 18:172–173.

8. Fontana C, Favaro M, Pelliccioni M, Pistoia ES, Favalli C. Use of the MicroSeq 500 16S rRNA gene-based sequencing for identification of bacterial isolates that commercial automated systems failed to identify correctly. J Clin Microbiol. 2005; 43:615–619.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download