Abstract
Background
Hepatitis E virus (HEV) causes epidemics in developing countries and is primarily transmitted through the fecal-oral route. There have been recent reports on the zoonotic spread of the virus, and several animal species, primarily pigs, have been recognized as reservoirs of HEV. Because of its possible spread, there is an urgent need of a method for the cost-effective production of HEV proteins that can be used as diagnostic antigens for the serological detection of anti-HEV antibodies.
Methods
The HEV open reading frame (ORF)2 protein was purified from plant tissue by using immobilized metal-anion chromatography (IMAC). The recombinant protein was used to develop an in-house ELISA for testing anti-HEV antibodies in both human and swine sera. Thirty-six serum samples collected from patients with serologically proven HEV infection with commercial kits were tested for anti-HEV IgG antibodies by using the plant-expressed protein. Forty-five serum samples collected from apparently healthy pigs in Bulgarian farms were also tested.
Results
We confirmed the transient expression and purification of a truncated version of the HEV genotype 3 capsid protein in Nicotiana benthamiana and its usefulness as a diagnostic antigen. ELISA showed the presence of anti-HEV IgG antibodies in 29 of the 36 human samples. The in-house ELISA showed anti-HEV IgG antibodies in 34 of the 45 pigs.
Hepatitis E virus (HEV) is the causative agent of epidemic and sporadic viral hepatitis, which has become a major public health concern in both developing and developed regions. Sporadic cases of zoonotic and food-borne infection have been reported in both HEV endemic and non-endemic areas [1]. Several investigations have shown that domestic pigs in industrialized areas are widely infected with HEV genotype 3, and autochthonous cases of hepatitis E have been reported [23]. The mortality rate of HEV infection ranges from 1% to 4%, and can be as high as 25% in pregnant women [4]. HEV is classified in the Hepeviridae family with at least four genotypes (1-4) of the virus infecting humans [5]. Genotypes 1 and 2 are restricted to humans, while genotypes 3 and 4 are zoonotic and responsible for autochthonous infections in humans [1]. HEV-3 is now considered an emerging pathogen, and is the most common genotype detected in both humans and swine in industrialized nations [267]. Several lines of evidence indicate the occurrence of the zoonotic transmission from pigs, wild boar, and deer to humans [89]. Furthermore, numerous reports have revealed the similarities in HEV-3 sequences detected in swine and humans from the same geographic are [1011]. In Europe, HEV-3 infections in pig farms are widespread with a prevalence of anti-HEV antibodies in pigs reaching 98%, and typically occur at the age of 2-6 months [12]. HEV infection in pig farms is also widespread in the USA [13]. However, to date, there is no information on the circulation of HEV in pigs and other animals in Bulgaria. HEV is not diagnosed routinely in pigs because the infection is often asymptomatic and is not associated with defects in growth [14]. This fact, together with sporadic cases of HEV infection occurring after the consumption of undercooked animal meat [151617], makes HEV a significant zoonotic disease that needs to be properly diagnosed.
HEV is a small, spherical, non-enveloped, single-stranded RNA virus [18]. The HEV genome has three open reading frames (ORFs). ORF2 encodes the capsid protein, which shows strong immunogenicity, and the antibodies generated in response to infection can efficaciously neutralize the virus [19]. Thus, the ORF2 capsid protein is an appropriate candidate for the serological diagnosis of HEV [202122].
Definitive diagnosis of HEV infection is based on detection of anti-HEV IgM antibodies or HEV RNA in serum samples. The presence of anti-HEV IgG is typically used to determine the seroprevalence in a population; however, both IgM and IgG antibodies are produced in people with acute HEV infection prior to the manifestation of clinical symptoms, which is potentially diagnostically relevant [23]. Therefore, patients presenting with infection symptoms will likely be positive for both anti-HEV IgM and IgG antibodies, although IgG is less indicative of an acute infection. Although several diagnostic kits for anti-HEV antibodies are available, they often provide contradictory results and are quite expensive [24]. Thus, the cost-effective production of recombinant proteins as diagnostic antigens is needed for the serological diagnosis of HEV. Although good expression has been achieved in both mammalian and bacterial cells, the process is quite expensive and cumbersome [25]. Therefore, plants may be a novel source for the cost-effective production of recombinant proteins useful for immunological studies. Plants have been explored as platforms for the production of diagnostic and therapeutic recombinant proteins owing to their scalability, safety, and their ability to perform eukaryotic post-translational modification [2526].
In this study, we developed a cost-effective and simple method for production of the HEV-3 ORF2 protein in Nicotiana benthamiana leaves, and demonstrated its usefulness for the serological detection of anti-HEV antibodies in both humans and swine. Transient expression in plant leaves enabled the efficient production of valuable proteins within one week. For this purpose, we used the Cowpea Mosaic Virus (CPMV)-based vector pEAQ-HT [27] for the expression of the HEV ORF2 protein in plant tissue. The expressed protein was recognized by both human and swine sera and can be used as a diagnostic reagent for the detection of anti-HEV IgG antibodies.
The nucleotide sequence of the swine genotype 3 HEV capsid protein (GenBank accession number DQ079627.1) lacking the first 109 amino acids (aa) from the N-terminus and 50 aa from the C-terminus (HEV 110-610) was synthesized by Life Technologies (Carlsbad, CA, USA). To maximize the levels of expression, the HEV ORF2 gene was codon-optimized for expression in N. benthamiana.
DNA fragments for insertion of the 6x His-tag at either the N- or C-terminus of HEV 110-610 was amplified by PCR. The primer pair HEV/C-end His-tag/Fw (AAATACCGGTATGGCTACTTCTCCT) and HEV/C-end His-tag/Rv (AGGCCTCGAGCTAATGATGGTGATGGTGATGAGCAAGAGCAGAGTGAGGAGCAAG) was used to clone the His-tag at the C-terminus, and the primer pair HEV/N-end His-tag/Fw (AAATACCGGTATGCATCACCATCACCATCATGCTACTTCTCCTGCTCCAGATACTGCT) and HEV/N-end His-tag/Rv (GGCCTCGAGCTAAGCAAGAG) was used for introducing a His-tag at the N-terminus. The PCR fragments were flanked by AgeI and XhoI restriction sites (New England Biolabs, Ipswich, MA, USA), which were used for the cloning of PCR fragments into the pEAQ-HT vector digested with the same enzymes. Finally, the plasmid DNA was purified and confirmed by PCR and sequencing. The E. coli XL1 blue strain was used for all cloning experiments.
The constructs were transformed into the electrocompetent A. tumefaciens strain LBA4404. After electroporation at 2.5 kV, Super Optimal Broth with catabolite repression (SOC) medium was immediately added, and the cells were left to recover for 1 hr at 28℃. The cells were then plated on LB agar containing 50 µg/mL rifampicin and 50 µg/mL kanamycin.
N. benthamiana plants were grown in glasshouses maintained at 25℃ and watered daily. Supplemental lighting was provided to maintain 16 hr of daylight in the winter months. Plants that were 3–4 weeks old were used for the transient expression experiments.
Inoculated liquid cultures of A. tumefaciens strain LBA4404 were grown at 28℃ in a shaking incubator for 24 hr in Luria-Bertani (LB) medium containing rifampicin and kanamycin. N. benthamiana leaves were agroinfiltrated via a syringe at an optical density at 600 nm of 0.4.
Leaf tissue was harvested at six days post infiltration (dpi). Large-scale sampling was conducted by removing any large veins and non-infiltrated tissue, and recording the leaf sample weight.
Samples were extracted by adding 3 volumes of the extraction buffer (phosphate buffered saline, PBS, pH 7.4) with complete EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics GmbH, Mannheim, Germany) and homogenized in a blender. Large cell debris was removed by squeezing the homogenate through one layer of Miracloth (Merck KGaA, Darmstadt, Germany). NuPAGE Bis-Tris Mini gels of 4–12% or 12% (w/v) acrylamide (Invitrogen, Carlsbad, CA, USA) and the protein pre-stained standard SeeBluePlus 2 (Invitrogen) or Spectra multicolor broad range protein ladder (TermoFisher scientific, Waltham, MA, USA) were used throughout the experiments.
The recombinant protein from leaf extracts was purified using immobilized metal-anion chromatography (IMAC) on a Ni-NTA column, according to the manufacturer's instructions (Qiagen, Hilden, Germany). Briefly, infiltrated leaf tissue was blended with binding buffer containing 10mM imidazole at 3-times the fresh weight of the leaves (FWT). The extract was filtered through two layers of Miracloth, and after centrifugation, the clarified extracts were added onto a Qiagen Ni-NTA column. The column was washed with 20mM imidazole and was eluted with a total of 4 mL of elution buffer containing 250mM imidazole. Protein concentrations were determined by densitometry of the stained gels using known amounts of purified bovine serum albumin as a control and by measuring the absorbance at 280 nm. Both proteins were purified up to 100 µg/g FWT using IMAC and detected by using SDS-PAGE and western blot.
The purified proteins were transferred from the SDS-PAGE gel onto a nitrocellulose membrane (Bio-Rad Laboratories Ltd, Hertfordshire, UK). Membranes were blocked with 5% (w/v) non-fat dried milk in PBS with 0.05% Tween-20 (v/v) (PBST) and incubated in the primary monoclonal antibody mouse anti-hepatitis E ORF2 antigen antibody (ab101124; Abcam, Cambridge, UK) diluted 1:3,000 at room temperature for 1 hr and washed with PBST. The bound antibody was detected with secondary anti-mouse antibody-horseradish peroxidase (HRP) (ThermoFisher Scientific) diluted 1:30,000. The emitted luminescence from the ECL detection reagents (GE Healthcare Life Sciences, Buckinghamshire, UK) was detected with the ImageQuant LAS 500 system (GE Healthcare Life Sciences). For western blot using human and swine serum as the primary antibodies, the membrane was incubated in human or swine serum (dilution 1:400), and bound antibodies were detected with secondary anti-human antibody-AP (Sigma-Aldrich, S.Louis, MO, USA) diluted 1:5,000 or secondary anti-pig antibody-AP (KPL, Gaithersburg, MD, USA) diluted 1:10,000, respectively. One-step NBT/BCIP substrate (ThermoFisher Scientific) was used. Western blot is universally accepted approving assay for detection of antibodies and it is considered a gold standard for affirmation of results. In this work, western blot was used to evaluate the quality of our in-house ELISA, by determining the rate of false-positives or false-negatives.
All human serum samples, with informed consents from the patients, were provided by the National Centre of Infectious and Parasitic Diseases, Sofia, Bulgaria. The serum samples were tested with commercial kits from Dia.Pro (Dia.Pro Diagnostic Bioprobes Srl, Milan, Italy), which detects serum IgG and IgM. All human sera were positive for both antibodies.
Thirty serum samples were collected from apparently healthy pigs (six months old) using blood collected from the ear vein by veterinarians. Additional 15 serum samples were collected from the intracardiac clot of slaughtered pigs at slaughterhouses post-mortem. Sera were aliquoted and stored at −20℃ until use. All swine serum samples were donated by the Department of Animal Genetics, Faculty of Veterinary Medicine, Trakia University, Bulgaria.
Microtiter plates (Greiner 96-well flat bottom) were coated with 50 µL/well of serial dilutions of purified protein in duplicate wells (12.5 to 100 ng/well) in PBS (pH 7.4) and incubated overnight at 4℃. After three washes with PBST, plates were incubated with 200 µL/well of blocking solution (PBST–5% [w/v] dry milk) and incubated for 1 hr at room temperature. Serum diluted 1:100 in blocking buffer was then added.
Plates were washed again with PBST before the addition of 50 µL/well of an HRP-conjugated goat anti-human IgG or anti-swine secondary antibody diluted 1:10,000 (KPL) or 1:8,000 (ThermoFisher Scientific), respectively. After incubation with the secondary antibody, plate wells were washed three times before 50 µL/well of the substrate solution (o-phenylenediamine, Sigma-Aldrich) was added. Plates were incubated in the dark at room temperature from 10 to 20 min, and then the reaction was stopped by the addition of 50 µL/well of 1M H2SO4, and the plates were read at 492 nm in a plate reader Epoch Microplate Spectrophotometer (BioTek Instruments Inc., Winooski, VT, USA). Ten previously characterized negative human serum samples by using commercial kits from Dia.Pro (Dia.Pro Diagnostic Bioprobes Srl) and four negative swine serum samples by using porcine kit PrioCheck HEV Ab (ThermoFisher Scientific) were used as controls to determine the cut-off value for the human and swine anti-HEV IgG ELISA assay. The cut-off value was calculated as absorbance value of the test sample/absorbance value of the negative control or P/N of ≥2.5. Samples with absorbance values above the cut-off (P/N) were considered positive [20].
A synthetic gene of the ORF2 of HEV lacking the first 109 aa at the N-terminus and the last 50 aa at the C-terminus was designed. The sequences were codon-optimized for the N. benthamiana genome to increase the yields of expressed protein [28]. The synthetic gene was used as template for insertion of a 6x His-tag at either the N- or C-terminus by the PCR DNA amplification method (see Methods). The resultant PCR products were cloned into pEAQ-HT and, after confirming the sequence of the insert, the constructs were transformed into A. tumefaciens, and N. benthamiana leaves were agroinfiltrated.
Plant leaves infiltrated with HEV 110-610 C-end expressed a large amount of a 54.6 kDa protein as revealed by SDS-PAGE followed by staining with Instant Blue (Fig. 1A). Western blot analysis using anti-HEV ORF2 antibody confirmed that the plants successfully produced HEV 110-610 C-end protein (Fig. 1B). The expressed HEV 110-610 with His-tag at the N-terminus was also analysed by SDS-PAGE (Fig. 1C) and confirmed with western blot (Fig. 1D). The 54.6 kDa band was absent in extracts from tissues infiltrated only with pEAQ-HT (data not shown).
The purified protein was also run on SDS-PAGE gels and detected with anti-HEV IgG-positive swine sera (Fig. 2A) and with anti-HEV IgG-positive human sera (Fig. 2B). In addition, the samples were detected with anti-HEV ORF2 antibody as a control (Fig. 2C). Both the human and swine sera recognized a protein band of 54.6 kDa (arrows in Fig. 2) and some aggregates and degradation products.
The purified HEV 110-610 C-end and N-end His-tag proteins reacted similarly to human sera previously characterized as positive for anti-HEV antibodies. After obtaining the preliminary results with both proteins, we chose to perform all ELISA tests with the HEV 110-610 C-end His-tag protein. Five previously characterized negative human sera and five positive human sera were pooled and used for optimization of antigen concentration and serum dilution. Different concentrations of the HEV 110-610 recombinant protein were used. The optimal serum dilution was determined to be 1/100, and the antigen concentration was 50 ng/well (see Supplemental Data Fig. S1 and Fig. S2).
Thirty-six previously characterized positive human serum samples were tested by ELISA using the HEV 110-610 C-end His-tag protein as the antigen. The in-house ELISA showed anti-HEV IgG antibodies in 29 of the 36 human serum samples (Fig. 3).
Since western blot is more specific and confirmatory than ELISA, we also performed western blots with all of the human sera. All serum samples showed a positive signal on western blots. The sensitivity of the in-house ELISA assay was 80.5% compared with the commercial kit. From these data, we concluded that the plant-produced HEV 110-610-based ELISA test resulted in seven false-negative results (19.5%) (Fig. 4).
A total of 45 serum samples collected from healthy pigs in two Bulgarian farms and one slaughterhouse were tested for anti-HEV IgG antibodies. For specific HEV antibody detection in swine, we used both the plant-produced HEV 110-610-based ELISA and western blot. Four previously characterized negative swine sera and four positive swine sera were pooled and used for optimization of the antigen concentration and serum dilution. The optimal serum dilution was determined at 1/100, and the antigen concentration was 50 ng/well (see Supplemental Data Fig. S3).
The in-house ELISA showed antibodies in 34 of 45 pigs, indicating that 75.5% were positive for anti-HEV IgG antibodies (Fig. 3).
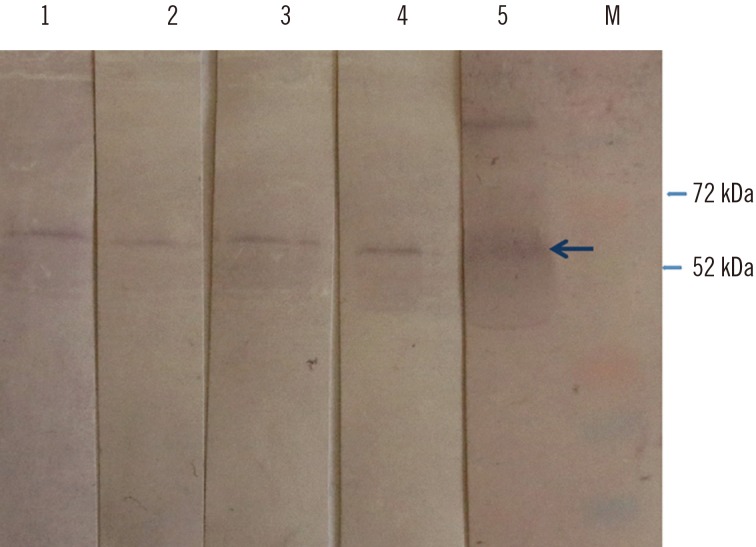
We then performed western blot with the 11 swine serum samples that scored negative by the ORF2-based ELISA. Four of these 11 samples tested positive by western blot (Fig. 5). This shows that the ELISA test resulted in four false-negative results or a rate of 8%.
We provide the first demonstration of the transient expression of truncated HEV 110-610 His-tagged protein in the plant N. benthamiana. The specificity of the expressed protein was confirmed by western blot using well-characterized monoclonal antibody, as well as human and pig sera. The optimal concentration of the antigen and serum dilution were assessed for an in-house ELISA for both swine and human sera. We demonstrated that the plant-produced truncated HEV ORF2 protein used as a coating antigen in our in-house ELISA was able to detect anti-HEV IgG antibodies in human sera with similar accuracy compared with that of the commercial kit. Since western blot is considered the gold standard, we used this method to confirm the ELISA-negative results. Anti-HEV IgG was detected in all human serum samples tested by the western blot. It is possible that some linear epitopes have been unmasked compared with the native antigen used in the ELISA. Furthermore, using the same method described herein, the recombinant plant ORF2 can also be used for the detection of serum IgM, which is considered to be more critical for the diagnosis of acute HEV infection (data not shown). Moreover, we could confirm the presence of the anti-HEV IgG antibodies in pigs in Bulgaria. Because pigs ordinarily are not tested for HEV infection, additional testing needs to be done to assess the seroprevalence of anti-HEV antibodies in Bulgarian farms.
Plant leaves infiltrated with pEAQ-HT HEV 110-610 with a His-tag at either the N- or C-terminus resulted in the expression of a 54.6 kDa protein at up to 100 µg/g of FWT. For instance, it has been reported that 1 mg of ORF2 protein can be produced per liter of E. coli culture [29]. Using the same protocol, 1 L of culture would produce enough protein to process 10,000 test samples, while 1 kg of plant tissue could produce enough protein to process 1,000,000 test samples at the yield described herein. This implies that one would need 100 L of bacterial culture to process the same number of samples as could be processed with the protein produced by 1 kg of plant tissue, which is much easier to obtain and much more cost-effective. If the production capacity is scaled up to levels used in the commercial production of plant-derived proteins (i.e., hundreds of kilos), then the final cost of the diagnostic reagent can quickly be lowered, improving its availability to those who need to administer the tests.
In conclusion, the HEV 110-610 recombinant protein can be produced in high yields at a relatively low cost in plant tissue, and could be used as a diagnostic antigen in the ELISA-based detection of anti-HEV antibodies in both human and swine sera.
Acknowledgments
The authors' research on HEV is supported by grants from the Bulgarian Science Fund project DMU03/33, FEBS Collaborative Experimental Scholarship for Central and Eastern Europe, and the UK Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004596/1).
References
1. Meng XJ. Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis. 2013; 33:41–49. PMID: 23564388.
2. Di Bartolo I, Ponterio E, Castellini L, Ostanello F, Ruggeri FM. Viral and antibody HEV prevalence in swine at slaughterhouse in Italy. Vet Microbiol. 2011; 149:330–338. PMID: 21216541.
3. Colson P, Romanet P, Moal V, Borentain P, Purgus R, Benezech A, et al. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg Infect Dis. 2012; 18:1361–1364. PMID: 22840196.
4. Kumar A, Beniwal M, Kar P, Sharma JB, Murthy NS. Hepatitis E in pregnancy. Int J Gynaecol Obstet. 2004; 85:240–244. PMID: 15145258.
5. Smith DB, Purdy MA, Simmonds P. Genetic variability and the classification of hepatitis E virus. J Virol. 2013; 87:4161–4169. PMID: 23388713.
6. De Silva AN, Muddu AK, Iredale JP, Sheron N, Khakoo SI, Pelosi E. Unexpectedly high incidence of indigenous acute hepatitis E within South Hampshire: time for routine testing? J Med Virol. 2008; 80:283–288. PMID: 18098134.
7. Pavio N, Meng XJ, Renou C. Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res. 2010; 41:46. PMID: 20359452.
8. Berto A, Martelli F, Grierson S, Banks M. Hepatitis E virus in pork food chain, United Kingdom, 2009–2010. Emerg Infect Dis. 2012; 18:1358–1360. PMID: 22840183.
9. Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008; 48:494–503. PMID: 18192058.
10. van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, et al. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg Infect Dis. 2001; 7:970–976. PMID: 11747723.
11. Takahashi K, Kitajima N, Abe N, Mishiro S. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology. 2004; 330:501–505. PMID: 15567444.
12. Meng XJ. Recent advances in Hepatitis E virus. J Viral Hepat. 2010; 17:153–161. PMID: 20040046.
13. Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002; 40:1326–1332. PMID: 11923352.
14. Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Nat Acad Sci USA. 1997; 94:9860–9865. PMID: 9275216.
15. Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011; 161:23–30. PMID: 21316404.
16. Pavio N, Merbah T, Thébault A. Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg Infect Dis. 2014; 20:1925–1927. PMID: 25340373.
17. Mizuo H, Yazaki Y, Sugawara K, Tsuda F, Takahashi M, Nishizawa T, et al. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J Med Virol. 2005; 76:341–349. PMID: 15902701.
18. Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991; 185:120–131. PMID: 1926770.
19. Meng J, Dai X, Chang JC, Lopareva E, Pillot J, Fields HA, et al. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology. 2001; 288:203–211. PMID: 11601892.
20. Jiménez de Oya N, Galindo I, Gironés O, Duizer E, Escribano JM, Saiz JC. Serological immunoassay for detection of hepatitis E virus on the basis of genotype 3 open reading frame 2 recombinant proteins produced in Trichoplusia ni larvae. J Clin Microbiol. 2009; 47:3276–3282. PMID: 19656986.
21. Baechlein C, Meemken D, Pezzoni G, Engemann C, Grummer B. Expression of a truncated hepatitis E virus capsid protein in the protozoan organism Leishmania tarentolae and its application in a serological assay. J Virol Methods. 2013; 193:238–243. PMID: 23747546.
22. Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Nat Acad Sci USA. 2009; 106:12986–12991. PMID: 19620712.
23. Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med. 2012; 367:1237–1244. PMID: 23013075.
24. Bendall R, Ellis V, Ijaz S, Ali R, Dalton H. A comparison of two commercially available anti-HEV IgG kits and a re-evaluation of anti-HEV IgG seroprevalence data in developed countries. J Med Virol. 2010; 82:799–805. PMID: 20336757.
25. Mett V, Farrance CE, Green BJ, Yusibov V. Plants as biofactories. Biologicals. 2008; 36:354–358. PMID: 18938088.
26. Lomonossoff GP, D'Aoust MA. Plant-produced biopharmaceuticals: A case of technical developments driving clinical deployment. Science. 2016; 353:1237–1240. PMID: 27634524.
27. Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009; 7:682–693. PMID: 19627561.
28. Love AJ, Chapman SN, Matic S, Noris E, Lomonossoff GP, Taliansky M. In planta production of a candidate vaccine against bovine papillomavirus type 1. Planta. 2012; 236:1305–1313. PMID: 22718313.
29. Taherkhani R, Makvandi M, Farshadpour F. Development of enzyme-linked immunosorbent assays using 2 truncated ORF2 proteins for detection of IgG antibodies against hepatitis E virus. Ann Lab Med. 2014; 34:118–126. PMID: 24624347.
SUPPLEMENTARY MATERIALS
Supplemental Data Fig. 1
Optimization of antigen concentration. Previously characterized positive and negative human sera were tested by ELISA against serial dilutions (100 to 12.5 ng/well) of purified hepatitis E virus 110-610 C-end His-tag protein. Data in each panel are shown as the average absorbance of five negative and five positive human sera.
Supplemental Data Fig. 2
Optimization of human serum dilution. Different serial dilutions of human sera (1/50 to 1/400) were tested by ELISA against 50 ng of purified hepatitis E virus 110-610 C-end His-tag protein. Data are shown as the average absorbance of five negative and five positive human sera.
Supplemental Data Fig. 3
Optimization of swine serum dilution. Different serial dilutions of swine sera (1/50 to 1/400) were tested by an ELISA against 50 ng of purified hepatitis E virus 110-610 C-end His-tag protein. Data are shown as the average absorbance of four negative and four positive swine sera.
Fig. 1
(A) SDS-PAGE of expressed and purified hepatitis E virus (HEV) 110-610 C-end His-tag protein; (B) Western blot of the gel with anti-HEV ORF2 antibody; (C) SDS-PAGE of expressed and purified HEV 110-610 N-end His-tag protein; (D) Western blot of the gel with anti-HEV ORF2 antibody. M, protein marker; 1, crude extract; 2, supernatant; 3, pellet; 4, flow-through; 5, washing step; 6–9, elutes of the 54.6 kDa protein.
Abbreviations: SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; ORF2, open reading frame 2.
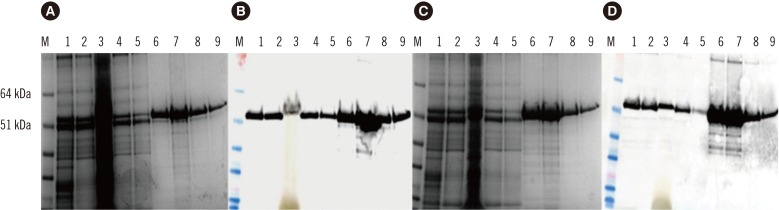
Fig. 2
Western blot of purified hepatitis E virus (HEV) 110-610 His-tag 54.6 kDa protein. Membranes were incubated with swine serum (A), human serum (B), and monoclonal antibody (anti-hepatitis ORF2 antibody) (C). M, protein marker; 1, HEV 110-610 N-end His-tag protein; 2, HEV 110-610 C-end His-tag protein (arrows).
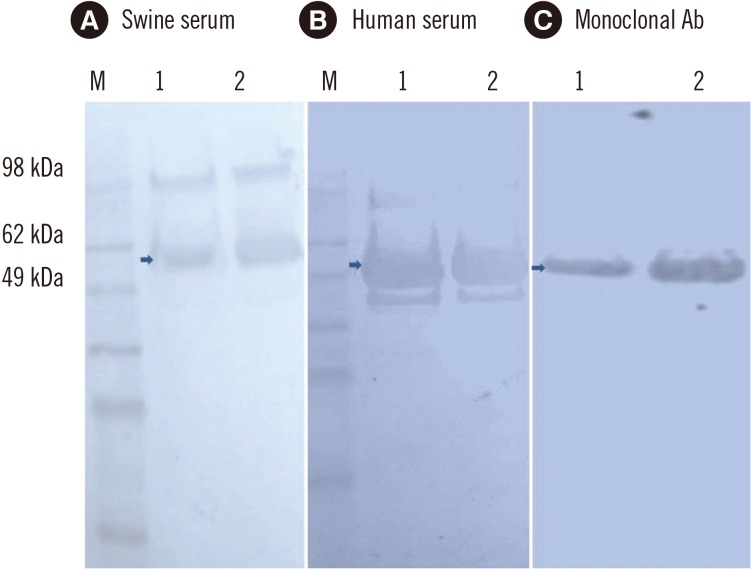
Fig. 3
In–house ELISA of serum samples based on the hepatitis E virus (HEV) 110-610 C-end His-tag protein. The cut-off value (CO) is indicated by a horizontal line. Samples were considered positive (in red), if the absorbance was above the cut-off value (P/N) (see Methods).
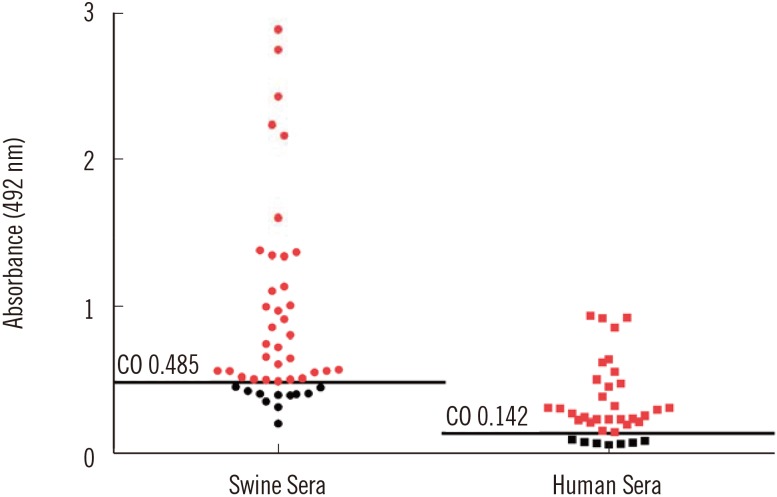
Fig. 4
Western blot probed with hepatitis E virus (HEV) 110-610-based ELISA negative human sera from this study (lanes 1-7). Arrow indicates the 54.6 kDa protein of interest. There are some aggregates and degraded products. Five HEV negative human sera previously characterized by commercial kit were used as control for western blot specificity (data not shown). M, protein marker.
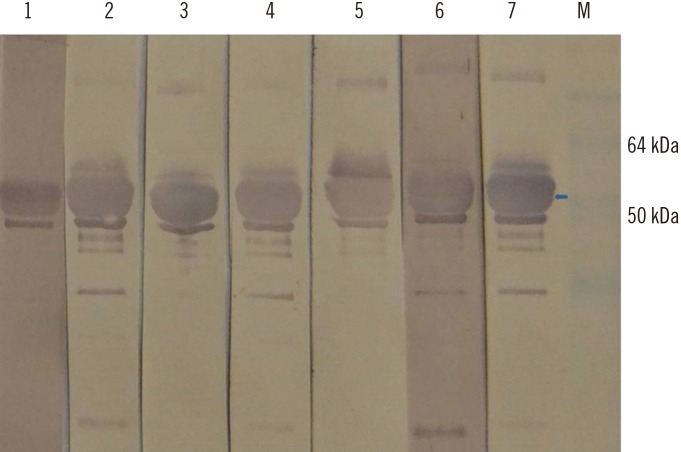
Fig. 5
Western blot of the hepatitis E virus (HEV) 110-610 C-end His-tag protein, probed with ELISA negative swine sera from this study (lanes 1–4), and with positive serum (lane 5). M, protein marker. Swine sera recognized a protein band of 54.6 kDa (arrow). Four HEV negative swine sera previously characterized by commercial kit were used as control for western blot specificity (data not shown).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download