Dear Editor,
AML is a group of heterogeneous diseases derived from various cytogenetic and molecular abnormalities that are significant for diagnosis, treatment, and prognosis in many cases. We report a rare case of AML harboring t(11;19)(p15;p12) that was detected with whole-genome sequencing (WGS).
A 33-yr-old woman was admitted to a local hospital with ab-dominal pain and diarrhea for five days. After conservative treatment, the symptoms improved. However, definite leukocytosis persisted; therefore, the patient visited our hospital for further evaluation. The initial complete blood count showed an Hb level of 7.6 g/dL, white blood cell (WBC) count of 29.48×109/L, and platelet count of 53×109/L. On the peripheral blood smear, differential counts revealed 15% segmented neutrophils, 16% lymphocytes, 60% monocytes, 1% eosinophils, 1% atypical lymphocytes, 1% promyelocytes, 6% blasts, and 4 nucleated red blood cells (RBCs) per 100 WBCs. RBCs were macrocytic and hyperchromic with mild polychromasia and anisocytosis. The bone marrow (BM) aspirate smear showed increased myeloblasts and monoblasts (7.6% and 42.0% of all nucleated cells, respectively) (Fig. 1A). Some blasts were positive for myeloper-oxidase (MPO), Sudan black B, specific esterase, and non-specific esterase stains, and showed fine granular positivity for periodic acid-Schiff stain. BM biopsy revealed a markedly hypercellular marrow (95–100% cellularity) with infiltrations of leukemic blasts.
Flow cytometric analysis revealed the blasts to be positive for CD11c, CD14, CD33, CD64, CD71, MPO, and HLA-DR, which indicated acute leukemia of the myeloid lineage with a monocytic component. Chromosomal analysis of the BM specimen showed 46,XX,t(11;19)(p15;p12)[26]/46,XX[1] (Fig. 1B). FISH analysis showed normal genotypes for AML1-ETO, PML-RARα, CBFB, and MLL. In the real-time allele-specific PCR assay for FLT3 mutations, a D835Y mutation was detected. Multiplex reverse transcriptase PCR using Hemavision kit (DNA Technology, Aarhus, Denmark) did not detect any other mutations.
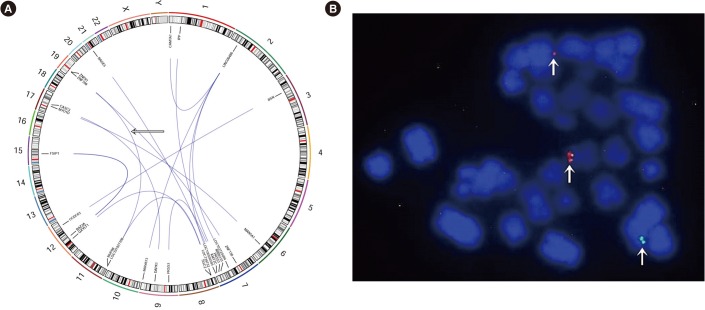
For further evaluation, we sent the BM samples out for WGS and RNA sequencing (RNA-seq) analysis. The WGS analysis was performed by using the HiSeq X Ten system (Illumina, San Diego, CA, USA), and RNA-seq analysis was performed by using the 100-bp paired-end mode of the TruSeq Rapid PE Cluster kit and the TruSeq Rapid SBS kit (Illumina, San Diego, CA, USA). The WGS analysis revealed the presence of 16 in-frame gene fusions, including a fusion between NUP98 and ZNF91 (Fig. 2A). To clarify these findings, additional FISH analyses using the NUP98 break-apart FISH probe kit (Empire Genomics, New York, NY, USA) were performed and revealed the presence of a break-apart rearrangement at chromosome 11p15 in 300 of 305 (98.4%) cells (Fig. 2B).
The patient was diagnosed as having acute myelomonocytic leukemia and was treated with idarubicin and cytarabine for induction chemotherapy. The blast count decreased gradually, and complete remission was achieved after reinduction chemotherapy. The follow-up chromosomal analysis of the BM specimen showed a normal karyotype.
To our knowledge, this is the first report of a case of AML harboring t(11;19)(p15;p12) in the world. From the WGS analysis with RNA expression data and a literature review, the fusion between NUP98 at chromosome 11p15.4 and ZNF91 at 19p12 resulted in a knockdown of both genes, which further dysregulated TP53 pathways [1]. The NUP98 gene encodes the major component protein of the nuclear pore complex that regulates the nucleocytoplasmic transport of proteins and mRNA [23]. Since the NUP98-HOXA9 fusion in AML harboring t(7;11)(p15; p15) was reported as the first case, more than 30 NUP98 partners have been identified in several hematologic diseases, including AML, MDS, CML in blast crisis, ALL, and bilineage/biphenotypic leukemia [456]. NUP98 fusion proteins have oncogenic properties, whereas NUP98 is a potential tumor suppressor and regulates the post-transcriptional expression of certain TP53 target genes [1].
In this case, the partner of the NUP98 gene was ZNF91, located on chromosome 19p12. However, in the RNA-seq analysis, no significant gene fusions were detected, and expression of the NUP98 gene decreased in the patient compared with data of the Cancer Genome Atlas (https://gdc.cancer.gov/). These results partially support that knockdown of the NUP98 gene by the gene–gene fusion probably played a role in the consequent defect of TP53 regulation in this case.
In conclusion, we report a first case of NUP98 rearrangement in AML with t(11;19)(p15;p12) in the world. More cases with similar karyotypes and genetic abnormalities should be evaluated to establish their effects on leukemogenesis.
References
1. Singer S, Zhao R, Barsotti AM, Ouwehand A, Fazollahi M, Coutavas E, et al. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell. 2012; 48:799–810. PMID: 23102701.
2. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002; 158:915–927. PMID: 12196509.
3. Gu BW, Wang Q, Wang JM, Xue YQ, Fang J, Wong KF, et al. Major form of NUP98/HOXC11 fusion in adult AML with t(11;12)(p15;q13) translocation exhibits aberrant trans-regulatory activity. Leukemia. 2003; 17:1858–1864. PMID: 12970787.
4. Borrow J, Shearman AM, Stanton VP Jr, Becher R, Collins T, Williams AJ, et al. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996; 12:159–167. PMID: 8563754.
5. Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996; 12:154–158. PMID: 8563753.
6. Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011; 118:6247–6257. PMID: 21948299.
Fig. 1
Findings of the bone marrow (BM) aspirate smear and Giemsa-banded karyogram. (A) Leukemic blasts with mixed myeloblasts and monoblasts on the BM aspirate smear (Wright-Giemsa stain, ×1,000). (B) Giemsa-banded karyogram of the BM cells at diagnosis: 46,XX, t(11;19)(p15;p12). The vertical arrows indicate abnormal chromosomes.
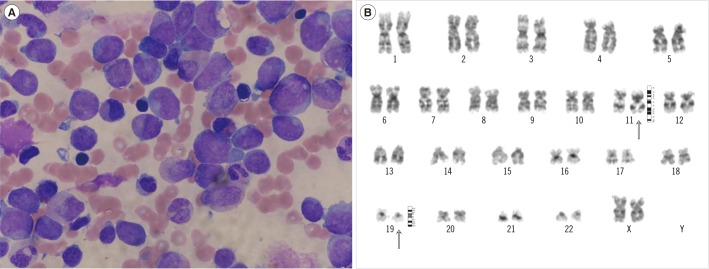
Fig. 2
The whole-genome sequencing (WGS) and FISH analyses. (A) Circos plot showing the in-frame gene fusions discovered in the WGS analysis. The lines traversing the ring indicate the gene fusions between each involved gene, and the outermost digits, X, and Y represent each chromosome number. The horizontal arrow points to the line indicating the fusion between NUP98 at chromosome 11p15.4 and ZNF91 at 19p12. (B) Metaphase FISH with the NUP98 break-apart probe. The vertical arrows indicate the presence of a break-apart rearrangement at chromosome 11p15.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download