Leptotrichia species are non-spore forming, anaerobic, gram-negative rods that exist as part of the normal flora of the human oral cavity and female genitalia. Systemic infections with
Leptotrichia species are rare and occur primarily in immunocompromised patients [
1]; infections including bacteremia [
2], periodontitis [
3], and infective endocarditis [
4] have been reported. Phenotypic identification methods have been unsuccessful in identifying these organisms to the species level. It is therefore important to assess the clinical and microbiological characteristics of
Leptotrichia species. The purpose of the present study was to investigate the microbiological characteristics of
Leptotrichia species in order to improve clinical laboratory identification.
We conducted a retrospective review of all 34,928 microorganisms isolated from blood cultures over a six-year period between January 2010 and May 2016. All isolates identified as Leptotrichia species were included and Gram stain and phenotypic and molecular identification results were reviewed. In addition, medical records were reviewed to obtain data regarding patient age, sex, underlying disease(s), presenting signs and symptoms, antimicrobial therapies, and clinical outcomes.
Blood samples were inoculated into aerobic and anaerobic blood culture bottles (bioMérieux, Marcy l'Etoile, France) and incubated for 5 days in a BacT/ALERT 3D blood culture instrument (bioMérieux). Isolates from aerobic blood cultures were subcultured on blood agar plates, MacConkey agar plates, and chocolate agar plates. Isolates from anaerobic blood cultures were subcultured on blood agar plates, MacConkey agar plates, and Brucella agar plates. Colony morphology and microscopic appearance were evaluated and conventional biochemical identification was performed using the VITEK 2 system (bioMérieux).
16S ribosomal RNA (rRNA) sequencing was used for molecular identification. DNA was extracted from the isolates by using the MagNa Pure LC Total Nucleic Acid Isolation Kit (Roche, Mannheim, Germany). 16S rRNA primers were designed according to the CLSI guideline MM18-A [
5]. Direct sequencing was performed by using the BigDye Terminator Cycle Sequencing Kit 3.1 (Applied Biosystems, Foster City, CA, USA) on an ABI prism 3730 Genetic analyzer (Applied Biosystems). Database comparison was conducted with the NCBI BLAST, EzTaxon, and BIBI databases [
6]. Identification was performed according to CLSI guideline MM18-A [
5].
Two commercially available matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems, VITEK MS (bioMérieux) and Bruker Biotyper (Bruker Daltonics, Bremen, Germany), were employed to analyze the stored isolates. Identification involved a direct, on-plate preparation method based on the manufacturers' instructions. The VITEK MS identification system compared the characteristics of the obtained spectra with those of the VITEK MS v2.0 database. A confidence value ≥ 60 with a unique spectrum of a single organism indicated good species-level identification. For the Bruker system, a MicroFlex LT mass spectrometer (Bruker Daltonics) was used for analysis. The spectra were analyzed by using Bruker Biotyper 3.1 software. Scores ≥2.000 indicated identification to the species level and scores between 1.700 and 1.999 indicated identification to the genus level.
A total of five
Leptotrichia isolates were identified from the blood samples of five patients collected over a six-year period. The characteristics of these patients are shown in
Supplementary Table 1. The mean age of the patients was 32.4 yr. All patients were diagnosed as having malignancies; three had hematologic malignancies, and two had solid cancers.
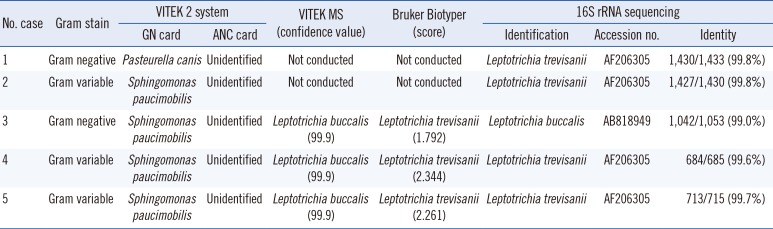
The phenotypic identification and sequencing results for the five isolates are shown in
Table 1. Primary subculturing resulted in the growth of pinpoint, gray, non-hemolytic colonies on blood agar plates following incubation at 35℃ with 5% CO
2 (
Fig. 1A, B); small gray colonies grew on the Brucella agar plates under anaerobic conditions at 37℃, while no growth was observed on the MacConkey agar plates. Gram stain analysis revealed long, fusiform and gram-negative or -variable bacilli (
Fig. 1C, D). On the basis of their morphology, these bacilli could be misidentified as
Fusobacterium species. Because
Leptotrichia species is not included in the VITEK 2 system (bioMérieux) database, identification of the isolates using the VITEK 2 system was unsuccessful. All five isolates were misidentified as
Pasteurella canis or
Sphingomonas paucimobilis by using the GN card and were not identified by using the ANC card. The characteristics of the isolates did not correspond with those of the identified organisms.
Fig. 1
Colony morphology and microscopy of Leptotrichia spp. Colony morphology of Leptotrichia buccalis isolated from case 3 (A) and Leptotrichia trevisanii isolated from case 5 (B). Gram stain of Leptotrichia species showing gram-negative (isolate from case 3) (C) and gram-variable (isolate from case 5) (D) cells (×1,000).
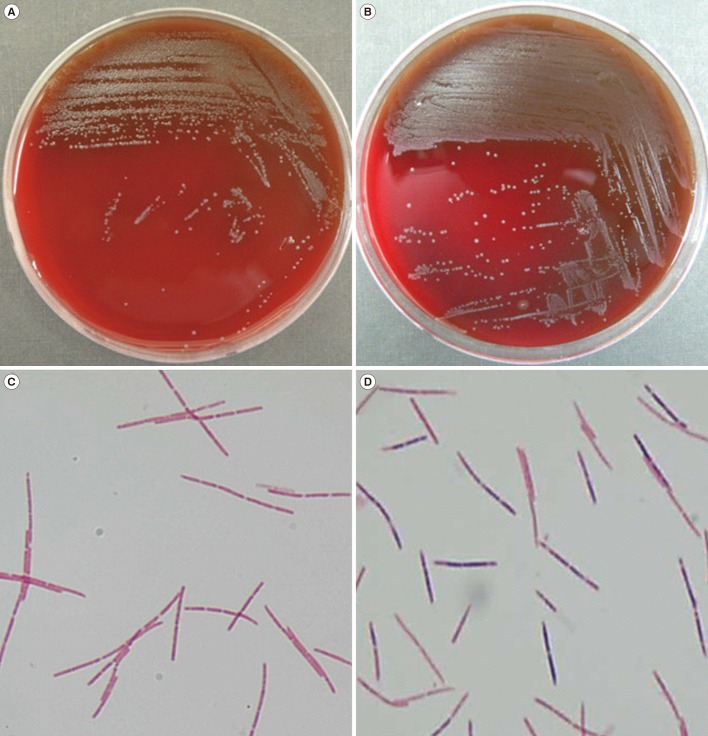

Table 1
Microbiological characteristics of the five Leptotrichia bacteremia cases
|
No. case |
Gram stain |
VITEK 2 system |
VITEK MS (confidence value) |
Bruker Biotyper (score) |
16S rRNA sequencing |
|
GN card |
ANC card |
Identification |
Accession no. |
Identity |
|
1 |
Gram negative |
Pasteurella canis
|
Unidentified |
Not conducted |
Not conducted |
Leptotrichia trevisanii
|
AF206305 |
1,430/1,433 (99.8%) |
|
2 |
Gram variable |
Sphingomonas paucimobilis
|
Unidentified |
Not conducted |
Not conducted |
Leptotrichia trevisanii
|
AF206305 |
1,427/1,430 (99.8%) |
|
3 |
Gram negative |
Sphingomonas paucimobilis
|
Unidentified |
Leptotrichia buccalis (99.9) |
Leptotrichia trevisanii (1.792) |
Leptotrichia buccalis
|
AB818949 |
1,042/1,053 (99.0%) |
|
4 |
Gram variable |
Sphingomonas paucimobilis
|
Unidentified |
Leptotrichia buccalis (99.9) |
Leptotrichia trevisanii (2.344) |
Leptotrichia trevisanii
|
AF206305 |
684/685 (99.6%) |
|
5 |
Gram variable |
Sphingomonas paucimobilis
|
Unidentified |
Leptotrichia buccalis (99.9) |
Leptotrichia trevisanii (2.261) |
Leptotrichia trevisanii
|
AF206305 |
713/715 (99.7%) |

A total of four isolates (cases 1, 2, 4, and 5) were identified as
Leptotrichia trevisanii by 16S rRNA gene sequencing. Over 99% homology was detected with
L. trevisanii type strain sequence AF206305. The next closest
Leptotrichia species sequences ranged from 96.0 to 97.0% similarity. The remaining isolate (case 3) was identified as
Leptotrichia buccalis, with 99% similarity to
L. buccalis reference strain AB818949. The second-matched sequence was identified as
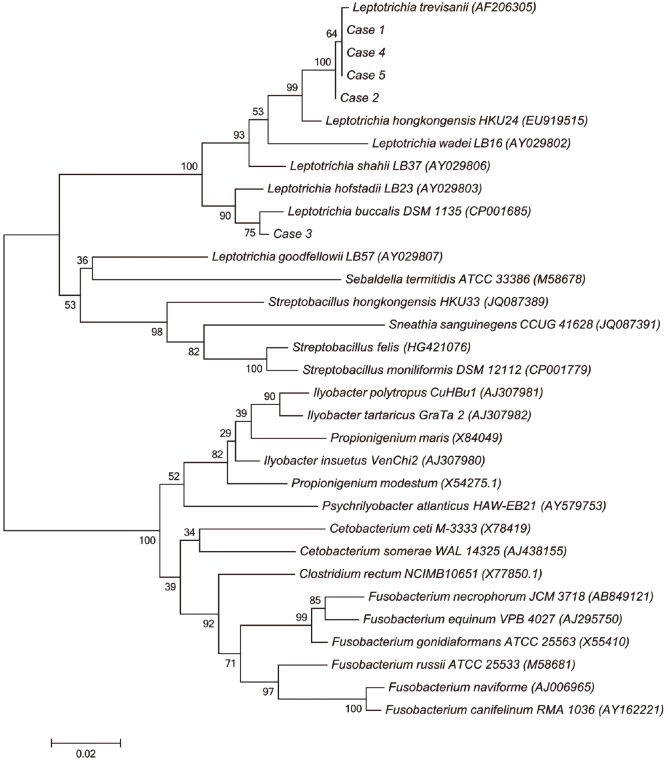
Leptotrichia hofstadii, with 97.8% similarity to the reference strain. A phylogenetic tree based on 1,000 bootstrap replicates of the 16S rRNA gene sequences was reconstructed with MEGA6 software (
http://www.megasoftware.net) (
Fig. 2).
Fig. 2
Neighbor-joining phylogenetic tree showing the relationships of the five isolates to related Leptotrichia species and other Fusobacteriaceae. Reference sequences are from species type strains; GenBank accession numbers are given in parentheses.

MALDI-TOF MS identification was available for only three recent isolates (cases 3-5). L. buccalis is the only species of this genus included in the VITEK MS database. L. buccalis (case 3) was correctly identified; however, two L. trevisanii isolates (cases 4 and 5) were misidentified as L. buccalis with a high confidence value of 99.9 by using VITEK MS. The Bruker Biotyper (Bruker Daltonics) database contains L. trevisanii and L. wadei. The two L. trevisanii isolates (cases 4 and 5) were correctly identified with scores ≥2.000. The case 3 isolate (L. buccalis) was identified as Leptotrichia sp. with a score of 1.792.
The genus
Leptotrichia is part of the family
Fusobacteriaceae. Currently, there are seven validly described
Leptotrichia species (
http://www.bacterio.net/leptotrichia.html):
L. buccalis,
L. goodfellowii,
L. hofstadii,
L. shahii,
L. trevisanii,
L. wadei, and
L. hongkongensis [
7].
Leptotrichia species are often misidentified as other species or not identified by using routine microbiological tests. While these species are anaerobic, pencil-shaped, gram-negative rods, they can appear as gram variable or even gram positive and grow under aerobic conditions, because they may retain crystal violet and have aerotolerant tendencies [
8]. These properties make identification of
Leptotrichia species more difficult. Because other species, such as
Actinomyces,
Arthrobacter,
Corynebacterium,
Mycobacterium,
Propionibacterium,
Bacillus,
Butyrivibrio, and
Clostridium, can exhibit gram-variable results [
9], microscopic analysis and colony morphology, together with a patient's clinical history, should be considered for accurate identification. Moreover, when gram-variable results are observed, the possibility of a mixed infection, older culture age, or technical errors should be considered.
In the present study, the automated VITEK 2 system could not identify Leptotrichia species. All five Leptotrichia species were misidentified as Pasteurella canis or Sphingomonas paucimobilis. Pasteurella canis is a member of the oropharyngeal flora in animals and causes zoonotic infection in humans. Thus, if a patient has no history of contact with animals, the possibility of misidentification should be considered. Misidentification of Leptotrichia species as an aerobe such as Sphingomonas paucimobilis can have a critical effect on clinical outcomes because routine antibiotics do not target anaerobes. Therefore, if the characteristics of an isolate do not correspond with those of an identified organism, MALTI-TOF MS or 16S rRNA sequencing should be considered for accurate identification. MALDI-TOF MS can successfully identify Leptotrichia species to the genus level. However, because of the limitations of the MALDI-TOF MS database, accurate identification of Leptotrichia to the species level is not possible. To date, only molecular methods using 16S rRNA sequencing were able to identify Leptotrichia species to the species level.
The incidence of anaerobic bacteremia is significantly higher in patients with hematologic malignancies [
10]. The most common source of anaerobic infection in these patients is the oropharynx. In non-hematologic patients, including patients with solid cancers, the most common source of anaerobic infection is the gastrointestinal tract. According to the previous study [
11], hematologic malignancy is the most common underlying disease in patients with
Leptotrichia bacteremia and the second most common underlying disease is solid cancer. Other characteristics of patients with
Leptotrichia bacteremia include neutropenia, a history of chemotherapy, and oropharyngeal mucosal damage. All patients in our study were immunocompromised patients with hematologic or non-hematologic malignancies. All patients were cured of infection by antibiotic treatment with negative follow-up blood culture results.
In conclusion, Leptotrichia species should be considered when gram-variable or -negative bacilli similar to Fusobacterium are observed. Identification based on biochemical testing is not possible; however, MALDI-TOF MS is a useful tool for identifying Leptotrichia species to the genus level. Molecular testing is required in order to identify Leptotrichia to the species level.






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download