INTRODUCTION
The detection and measurement of autoantibodies against nuclear and cytoplasmic antigens play an important role in the diagnosis of many autoimmune diseases such as systemic lupus erythematosus (SLE), mixed connective tissue diseases, rheumatoid arthritis, progressive systemic sclerosis, and chronic autoimmune hepatitis. The gold standard for antinuclear antibody (ANA) screening is indirect immunofluorescence (IIF) on human epithelial cells (HEp-2) [
12].
However, pattern assignment by manual fluorescence microscopic observation is time consuming and laborious. In addition, the interpretation could be subjective and conclusions can differ depending on operators. As a result, the requirement for automation and standardization of ANA testing has been highlighted. Currently, several automated systems for IF staining and interpretation have been introduced: AKLIDES (Medipan, Dahlewitz, Germany), EUROPattern (Euroimmun AG, Luebeck, Germany), HELIOS (Aesku Diagnostics, Wendelsheim, Germany), Image Navigator (Immuno Concepts, Sacramento, CA, USA), NOVA View (Inova Diagnostics, San Diego, CA, USA), and Zenit G-Sight (Menarini Diagnostics, Florence, Italy). Studies assessing the performance of these systems as an alternative to conventional manual microscopic interpretation have been reported [
345]. A previous study describing the parallel evaluation of the six currently available automated ANA-IIF systems showed that the overall sensitivity of all systems was 96.7% and the overall specificity was 89.2% for the discrimination between positive and negative signals, which was quite promising [
4]. However, relatively few studies have evaluated the usefulness of these automated systems by determining whether they can accurately recognize mixed patterns of ANA or less common patterns [
46]. EUROPattern Suite (Euroimmun AG, Luebeck, Germany), an automated system designed for computer-aided immunofluorescence microscopy (CAIFM) is composed of several hardware and software modules for fully automated image acquisition and evaluation, with regard to pattern recognition. Unlike other automated systems developed to recognize negative/positive results or simple patterns, the EUROPattern Suite software can assign variable mixed patterns on the basis of the software algorithm [
67].
The aim of this study was to evaluate the performance of EUROPattern Suite (Euroimmun AG, Luebeck, Germany) compared with conventional manual IIF microscopic interpretation for identifying both the presence of ANA and assigning the pattern of ANA.
Go to :

METHODS
1. Human sera
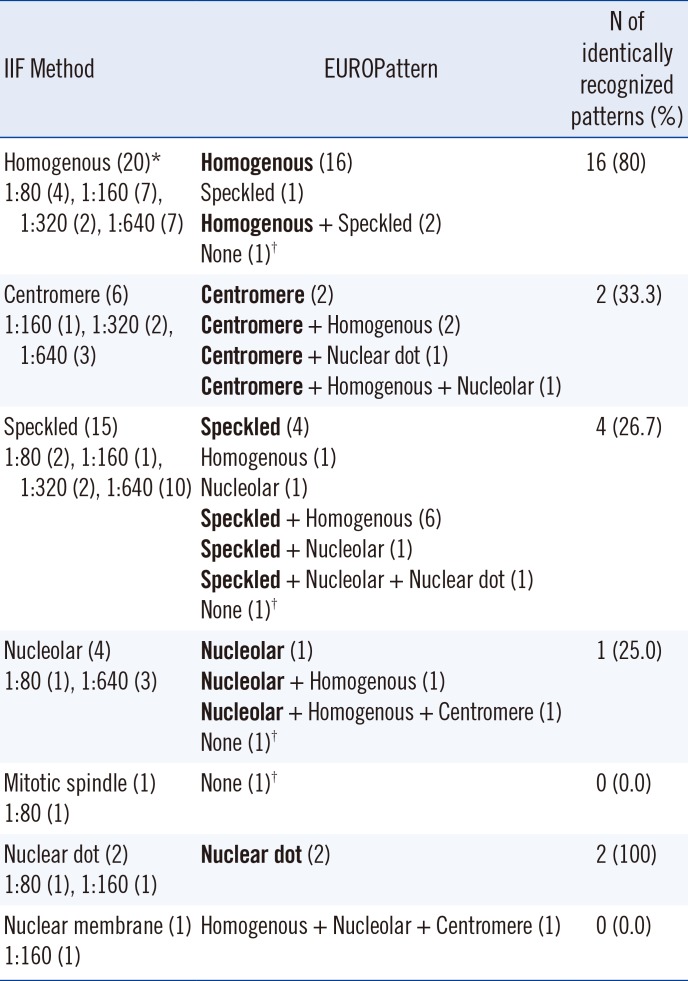
A total of 104 samples, including 70 ANA-positive sera and 34 ANA-negative sera, were collected from September to October 2015. Positive sera samples, which were tested by using the conventional indirect IIF assay, included samples with variable patterns with a titer of 1:80 to 1:640, which is comparable to 1+ and 4+, respectively. The specific patterns were assigned through manual IF microscopic observations by two experts; ANA-positive sera were divided into two groups: simple positive pattern (n=49) and mixed positive pattern (n=21). A simple pattern was defined as a single nuclear pattern and/or single cytoplasmic pattern. Twenty homogenous patterns (including eight dense fine speckled [DFS]), six centromere patterns, 15 speckled patterns, four nucleolar patterns, one mitotic spindle pattern, two nuclear dot patterns, and one nuclear membrane pattern were observed. A mixed pattern was defined as the presence of two or more nuclear patterns regardless of the existence of a cytoplasmic pattern. The patient diagnoses of 70 positive samples were categorized by reviewing patient medical records. Thirty-five patients (50%, 35/70) had systemic autoimmune diseases, including SLE (n=10), Sjogren syndrome (n=5), and systemic sclerosis (n=1), and 14 patients (20%, 14/70) were diagnosed as having organ specific autoimmune diseases such as autoimmune hepatitis. Twenty-one patients (30%, 21/70) could not be grouped into a particular category because they exhibited poorly defined symptoms or signs.
This study was conducted according to the guidelines of the Declaration of Helsinki, and all procedures involving human subjects were approved by the Institutional Review Board of Samsung Medical Center (IRB No: SMC-2015-10-187-002).
2. Indirect Immunofluorescence assay
The manual ANA assay was performed by using the IIF method with HEp-2 cells (FLUORO HEPANA TEST, MBL, Nagoya, Japan). The cells were fixed on substrate slides and incubated with diluted sera (1:40) for 20 min at room temperature. Following washing with phosphate buffered saline (PBS), each slide was stained with fluorescein isothiocyanate (FITC)-conjugated anti-human globulin. The slides were incubated for additional 20 min and then washed with PBS. The final step involved embedding with buffered polyvinyl alcohol for microscopic observation. For the quantitative assay, 1:80, 1:160, 1:320, and 1:640 serial dilutions of the serum samples were used.
Subsequent to slide preparation, two experts assigned the patterns and titers independently without reference to the other slides. The following nuclear and cytoplasmic patterns were reported: homogenous, speckled, nucleolar, DFS, nuclear dots, centromere, nuclear membrane, cytoplasmic, others (such as weak positive), and negative.
3. EUROPattern Suite
EUROPattern Suite (Euroimmun AG) is an automated system designed for computer-aided evaluation of fluorescence images using HEp-20-10 cells. This system consists of an automated microscope, the laboratory management software EUROLabOffice, and the pattern recognition software EUROPattern. Preparation of IIF slides can be automatically performed by using the IF Sprinter and interpretation procedures with EUROPattern suite following manual upload of the slides on the EUROPattern microscope. Using a reference database of over 5,000 images, a suggestive titer of >1:40 and single or mixed patterns can be analyzed [
6]. Seven different nuclear patterns can be recognized by this automated system: homogenous, nucleolar, centromere, nuclear dot, nuclear membrane, speckled, and mitotic spindle. However, this system does not have the ability to assign the DFS patterns derived from antibodies against the DFS70 antigen.
4. Definitions and statistics
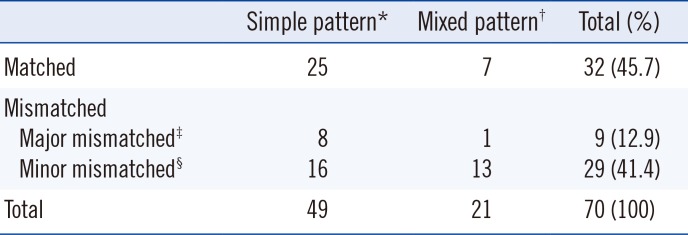
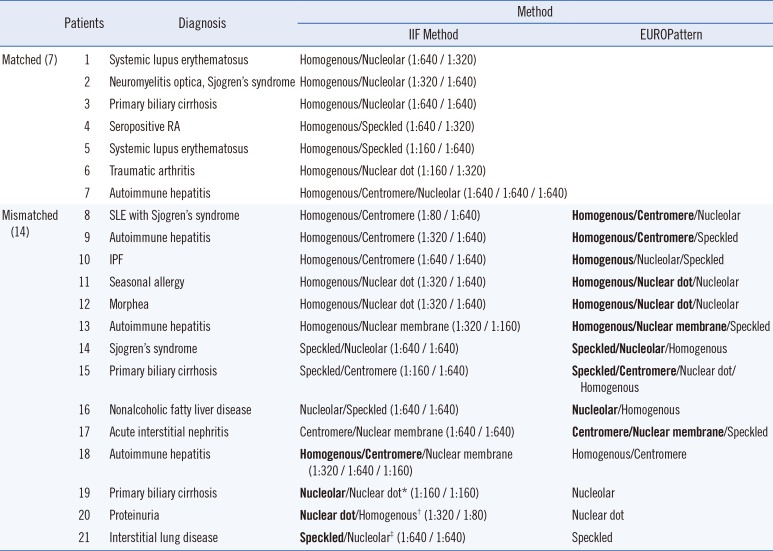
Sensitivity and specificity were calculated to evaluate the performance of EUROPattern Suite using the results of the manual IIF assay as a reference. In addition, the level of pattern recognition was classified as matched, major mismatched, and minor mismatched based on the manual IIF assay results. A matched designation indicates that all nuclear and cytoplasmic patterns were identical. A clinically informative pattern was considered as a major pattern; these included homogenous, nucleolar, speckled, and centromere patterns. On the basis of this definition, patterns were defined as major mismatched if one or more major pattern(s), including homogenous, nucleolar, speckled, and centromere patterns, were missing. Patterns were defined as minor mismatched if major patterns were assigned but either additional pattern(s) were present or minor patterns, such as nuclear dot or nuclear membrane, were missing.
The DFS pattern assigned by the IIF assay was considered a homogenous pattern, because the presence of the DFS70 antibody was not confirmed by enzyme immunoassay (EIA) method in this study.
We used the Cohen's kappa coefficient to measure interrater agreement between the manual IIF assay and the EUROPattern system for interpretation of the presence of ANA and the assignment of specific patterns. This kappa value was interpreted according to the following definition: >0.75, excellent; 0.40 to 0.75, fair to good; and <0.40, poor.
Go to :

DISCUSSION
In this study, we evaluated the performance of EUROPattern Suite for the detection of the presence of ANA and the assignment of simple and mixed patterns in samples positively identified by manual microscopic interpretation.
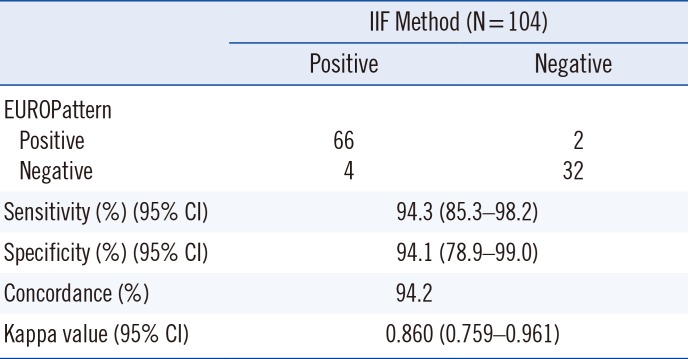
Considering the purpose of the ANA screening test and the higher proportion of negative samples in routine clinical settings, specificity and negative predictive value (NPV) are important factors. In our hospital, > 85% of requested samples were estimated as negative or weakly positive (data not shown). Although this study was not designed to estimate the NPV, we observed only four false negative results out of 70 randomly selected positive samples and two false positive results out of 34 randomly selected negative samples. These results suggest the potential applicability of EUROPattern Suite as a screening tool for ANA. The two falsely assigned patterns were cytoplasmic and centromere, and the assigned titers were all 1:160. Thus, the specificity of EUROPattern was 94.1%. All four samples that showed false negative results had relatively low titers (1:80). Particularly, one sample had a fluorescence pattern specific to the mitotic spindle apparatus (MSA). Typical autoantibodies for MSA, such as NuMA1 and NuMA2, are not commonly detected and may represent only 0.38% of ANAs [
8].
EUROPattern Suite and the manual microscopic assay showed concordance rate of 55.1% for simple patterns and 33.3% for mixed patterns, which appear to be insufficient from a practical view point. However, when considering the types of mismatched patterns, the concordance rate between both methods increased considerably to 83.7% and 95.2% for simple and mixed patterns, respectively. When we analyzed the prevalence of positive patterns in FANA over three months at our center, 25% of speckled patterns and 23% of homogenous patterns were observed among the positive patterns. In some studies, patients with strong FANA titers and homogenous or speckled patterns showed a higher prevalence of association with systemic rheumatoid diseases [
9]. Of our simple pattern results, 71.4% were homogenous or speckled patterns, which were the most prevalent and clinically correlated patterns.
Although the recognition rate for speckled patterns was low, most mismatched patterns were due to the assignment of additional patterns, not missing patterns. The recognition of further patterns in addition to major patterns may not be a critical failure for an automated system, although it necessitates additional processes, such as manual microscopic observation of slides or the verification of pictures taken by the automated system, to clarify more accurate ANA patterns. Importantly, homogenous pattern recognition was highly accurate not only for simple patterns but also for mixed patterns.
Although mixed patterns may comprise less than 10% of positive samples, patients with multiple patterns had an increased frequency of SLE and diseases of the scleroderma spectrum compared with patients with single FANA patterns [
10]. Consequently, the recognition of multiple FANA patterns would be helpful in providing diagnostic information to clinicians. However, in a routine diagnostic assay for detecting multiple ANA patterns, weak staining patterns can be obscured and potentially assigned as strong major patterns. In our mixed pattern analysis, only four cases of missed patterns were observed including nuclear dot and nuclear membrane patterns. Moreover, the major patterns that have strong intensity were all assigned perfectly.
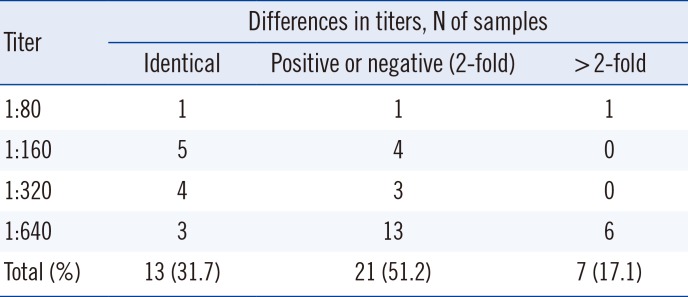
In addition to pattern recognition, we evaluated titer concordance between the two methods. Considering the subjective tendency of the conventional IIF assay, we postulated that a 2-fold difference in intensity was acceptable. Only seven samples exhibited >2-fold difference and had strong intensity (such as 1:640) by IIF assay. Therefore, the two methods were concordant in 82.9% (34/41) of samples, taking into account both patterns and titers. Furthermore, we found that this automated system detected more patterns with higher intensity compared with the conventional IIF assay.
Currently, all six commercial automated systems cannot identify DFS patterns. DFS patterns are related to the DFS70 antigen, which exists in healthy subjects and other inflammatory conditions such as interstitial cystitis, atopic dermatitis, and some types of cancer [
11]; therefore, their prevalence and significance in systemic autoimmune rheumatic diseases are relatively lower [
12]. Some studies have suggested that samples with a DFS staining pattern identified by IIF should be tested for anti-DFS70 antibodies, as well as the need for a test algorithm for DFS patterns [
1314]. In the current study, we considered DFS patterns as homogenous for the comparison of the two methods because the presence of the DFS70 antibody was not confirmed by a specific method.
Alongside the adoption of an automated image analyzer, one of the most important aspects that need to be considered is the standardization of ANA pattern nomenclature as well as the reporting format. Adoption of an automated staining system and image analyzer would reduce inter-laboratory variability; however, different pattern descriptions may hinder standardization. An international attempt to reach a consensus on the nomenclature for staining patterns and reporting results was made during the 12th International Workshop on Autoantibodies and Autoimmunity in 2014 [
1516]. Continuous concerted efforts are necessary to promote harmonization and understanding of ANA nomenclature and the standardization of ANA tests. In addition, it is possible for a reviewer to easily check the captured images with high resolution and the data can be stored for a long time without the need to retain the prepared slides. Recently, Mulliez et al [
17] reported one-year trial involving an automated ANA-IIF system, which resulted in important improvements with regard to workload and hands on time.
Our study has some limitations. The total number of study samples was relatively low, although a relatively high proportion of positive samples were included. Larger scope studies would be helpful to sufficiently evaluate the performance of EUROPattern Suite with a variety of patterns and clinical situations, which were not considered in this study. Although the detection of specific autoantibodies against extractable nuclear antigens (ENAs) plays a critical role in the diagnosis of autoimmune disease, we considered the identification of autoantibody specificity to be beyond the scope of the current study; therefore, we did not use these limited data to interpret our results.
In conclusion, this study observed good agreement between the manual and automated ANA IIF systems and demonstrated the potential benefits of automation when dealing with a large number of samples. The automated system can be used for the discrimination between positive and negative samples and the preparation of multiple slides in addition to serial dilution. Therefore, an automated system may increase laboratory efficiency and ensure standardization between immunologic laboratories. Furthermore, improving the accuracy of pattern recognition through the development of new algorithms may hasten the usage of these automated systems in many immunology laboratories [
18].
Go to :






 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download