INTRODUCTION
METHODS
1. Clinical yeast isolates
2. Yeast identification using the Bruker Biotyper system
3. Yeast identification using the VITEK MS system
4. Data and statistical analyses
RESULTS
1. Evaluation of the efficacy of the Biotyper system combined with different extraction methods
2. Performance comparisons between the Biotyper and VITEK MS systems
Table 1
Identification of 309 yeast isolates using the Biotyper and VITEK MS systems combined with different extraction methods, in comparison with the sequence-based identification
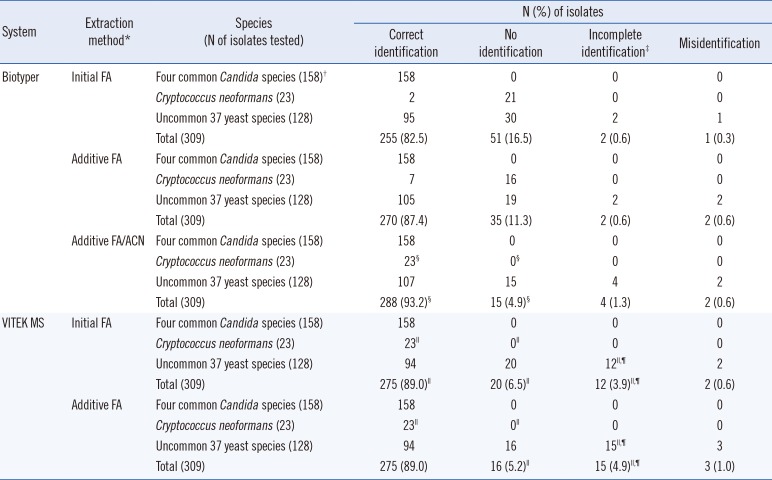
*Initial extraction was performed by using a simple formic acid (FA) method for all isolates (initial FA). The isolates that were not identified by using this method were retested by using the same FA extraction (both systems, additive FA) or an in-tube FA and acetonitrile method (Bruker Biotyper only, additive FA/ACN). The additive FA or FA/ACN results include the results of repeated tests, as well as the initial results of all acceptable identifications (scores ≥1.7).
†Including 57 Candida albicans, 47 Candida parapsilosis, 29 Candida tropicalis, and 25 Candida glabrata isolates.
‡Incomplete identification included (i) correct identification but a low level of discrimination group (two or three species were proposed, one of them was correct), (ii) identification of the species complex (e.g., the Candida parapsilosis or Candida haemulonii complexes), and (iii) correct identification of non-Candida yeasts to the genus level.
§P<0.05, between the results obtained with the Biotyper (initial FA vs additive FA or initial FA vs additive FA/ACN) within the same identification category.
∥P<0.05, between the results obtained with Biotyper or VITEK MS (both with initial FA or additive FA) within the same identification category.
¶P<0.05, between the results obtained with Biotyper preceded with additive FA/ACN and those obtained with the VITEK MS (initial FA or additive FA), within the same identification category.
3. Detailed identification of 42 yeast species using two MALDI-TOF systems
Table 2
Detailed identification of 309 isolates belonging to 42 yeast species using two MALDI-TOF MS systems in combination with additive formic acid extraction and in comparison with sequence-based identification
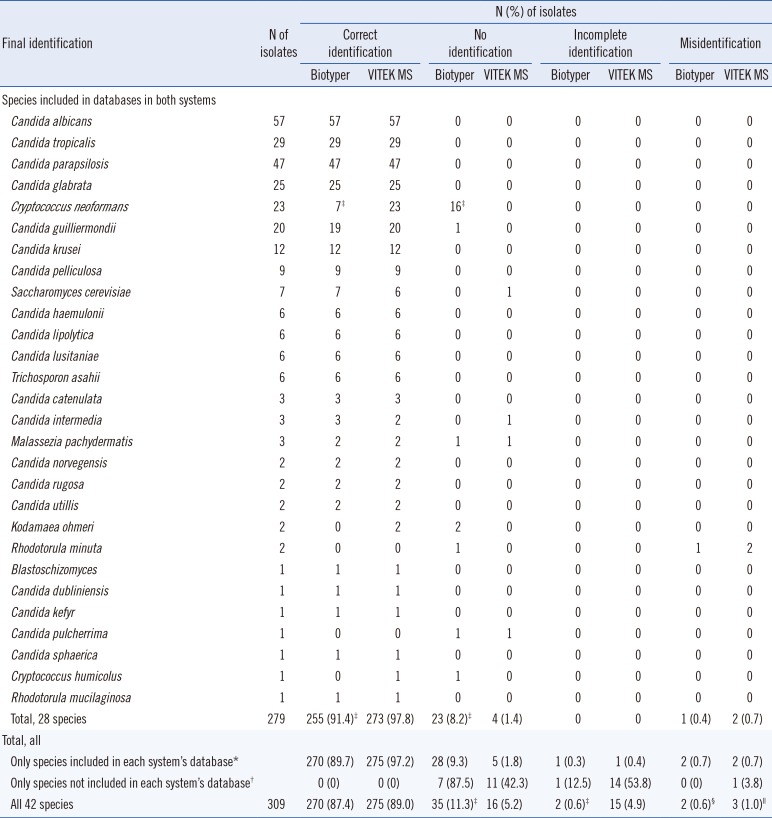
*Of 42 uncommon species, 37 (301 isolates) and 31 (283 isolates) species were included in the Biotyper and VITEK MS databases, respectively.
†Of 42 species, 5 (8 isolates) and 11 (26 isolates) species were not included in the Biotyper and VITEK databases, respectively.
‡P<0.05, between the results obtained with Biotyper and VITEK MS systems, within the same identification category.
§One isolate of Rhodotorula minuta and one isolate of Candida viswanathii were misidentified as Candida catenulate and Candida tropicalis, respectively, by the Biotyper.
∥Two isolates of Rhodotorula minuta were misidentified as Candida haemulonii and Candida catenulate, and one isolate of Candida metapsilosis was misidentified as Geotrichum capitatum by the VITEK MS.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download