Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) are important pathogens causing nosocomial infections in Korean hospitals. This study aimed to investigate the epidemiological and genetic diversity of clinical S. aureus isolates in healthcare settings from 2001 to 2008.
Methods
Samples and data were obtained from 986 individuals as part of the National Antimicrobial Surveillance Project, involving 10 regions nationwide. Molecular typing studies, including multilocus sequence typing (MLST) and staphylococcal cassette chromosome mec (SCCmec) typing were performed, and a representative clone of Korean MRSA was classified by combinational grouping using a DiversiLab (DL; bioMérieux, France) repetitive element polymerase chain reaction (rep-PCR) system.
Results
Nine Korean MRSA clones (KMRSA-1 to -9) were identified by analysis of genetic backgrounds and molecular characteristics. KMRSA-1 to -3, expressing clonal complex (CC) 5 (carrying SCCmec II), CC8 (carrying SCCmec III), and CC72 (carrying SCCmec IV) were spread nationwide. In contrast, KMRSA-6 was highly prevalent in Gyeongsangnam-do, and KMRSA-4 was highly prevalent in Jeollanam-do and Jeollabuk-do.
Conclusions
Epidemic KMRSA clones were genetically similar to major clones identified from the USA, with the exception of KMRSA-2, which had the SCCmec III type. Our results provide important insights into the distribution and molecular genetics of MRSA strains in Korea and may aid in the monitoring of MRSA spread throughout the country.
Staphylococcus aureus is an important human pathogen, and methicillin-resistant S. aureus (MRSA) has become a major cause of hospital- and community-acquired infection. Methicillin resistance rates in Korean hospitals have increased during recent decades, and MRSA reportedly accounts for 60-70% of all S. aureus isolates in tertiary hospitals in Korea. In 2009, the MRSA rate in Korea was reported to be as high as 81% in tertiary hospitals and 40% in non-tertiary hospitals [1].
In the United States of America (USA), Europe, and some parts of Asia, the incidence of MRSA has been monitored for prolonged periods to establish a national database of the multidrug-resistant clones distributed worldwide [2]. In the USA, USA300 and USA400 are the major clones frequently isolated in community-associated (CA) MRSA, and monitoring has provided a basis for the identification of toxic genes in these clones. Such monitoring allows for the recognition of outbreaks, and these clones provide a basis for monitoring the propagation and spread of domestic and international clones [3]. Monitoring efforts provide insights into the causes of increased resistance, which may be related to resistance obtained by various clones or to infection control measures, in which antibiotics select specific resistant bacteria. Additionally, monitoring tracks the rapid spread of bacteria among hospitals or between hospitals and the community, and helps evaluate the effects of increasing international travel and trade exchanges on the appearance of new resistant strains. Therefore, for effective analysis of the propagation of multidrug-resistant strains, the molecular epidemiological characteristics of domestic strains must be determined and compared with those of foreign strains.
Accordingly, the USA, Japan, Europe, and Denmark have established national organizations for compiling molecular epidemiologic information about multidrug-resistant strains of bacteria associated with infectious diseases, thereby allowing the monitoring of the prevalence of multidrug-resistant bacteria in the community or hospitals [456]. In Korea, specific analysis and monitoring have been performed only in some tertiary institutions at the national level, which makes the analysis of propagation patterns or trends difficult. Therefore, the identification of clones through the molecular genetic analysis of major resistant bacteria would improve our understanding of the propagation of MRSA and facilitate the establishment of disease management measures.
In this study, we identified major clones by targeting S. aureus isolated from medical care hospitals, including a geriatric hospital, in Korea. We analyzed the molecular genetic characteristics, classification, and characterization of MRSA isolates as well as the trends of national epidemic strains to identify resistant strains.
Samples were collected from hospitals that participated in the National Antimicrobial Surveillance Project; they were randomly and evenly selected from 10 districts (including Seoul, Gyeonggi-do/Incheon, Gangwon-do, Chungcheongbuk-do, Chungcheongnam-do/Daejeon, Gyeongsangbuk-do/Daegu/Ulsan, Gyeongsangnam-do/Busan, Jeollanam-do/Gwangju, Jeollabuk-do, and Jejudo) nationwide. A total of 986 S. aureus strains isolated from the surveillance cultures of clinical patients at general hospitals, hospitals and clinics, and geriatric hospitals between 2001 and 2008 were analyzed in this study. Most of the general hospitals were sentinel surveillance hospitals, and some isolates were identified by hospital-affiliated laboratories before the specimens were sent to Division of Antimicrobial Resistance, Korea National Institute of Health (KNIH).
Bacterial identification and antimicrobial susceptibility testing in this study were carried out with a VITEK 2 system (bioMérieux, La Balme les Grottes, France) and confirmed by the detection of mecA through PCR analysis. We used the following S. aureus strains: ATCC 25923, ATCC 29213, and ATCC 43300 (mecA, positive control) from ATCC (Manassas, VA, USA). If needed, antimicrobial susceptibility testing was performed with the disc diffusion method according to the CLSI guidelines [7]. S. aureus ATCC 29213 was used as a quality control strain for the determination of minimum inhibitory concentrations (MICs).
We analyzed data collected during an 8-yr period. Formal ethical approval for this study was obtained from Institutional Review Board of the Korea Centers for Disease Control and Prevention (KCDC). As part of this analysis, basic patient data were collected, including age (≥1 yr old), sex, and specimen type. Hospitals also provided consecutive, nonduplicate isolates for molecular typing. Cases were described as CA if MRSA was isolated from patients within 48 hrs of hospital admission or from patients who had none of the following risk factors: (i) history of hospitalization or surgery in the preceding calendar year, or (ii) residence in a long-term care facility (LTCF). Otherwise, they were considered to be healthcare-associated (HA) MRSA.
Staphylococcal cassette chromosome mec (SCCmec) types were determined with multiplex PCR [8]. PCR amplification for Panton-Valentine leukocidin (PVL)-encoding genes (lukS-PV and lukF-PV) was performed on representative isolates from each inferred from a multilocus sequence typing (MLST) clonal complex, as previously described [9].
For molecular typing, MLST and DiversiLab (DL; bioMérieux, Mercy l'Etoile, France) microbial typing procedures were performed as previously described [101112]. DL strain typing was determined via cluster analysis following the guidelines provided by the manufacturer. The isolates were categorized as indistin guishable, similar, or different according to bands interpreted using the DL analysis guide (software version 3.4).
For the strain typing of representative MRSA isolates, all MRSAs were categorized based on a combination of genetic differences determined through SCCmec typing and MLST, and the presence of PVL was also analyzed. For comparisons of the designated Korean MRSA (KMRSA) strains, the DL MRSA library database on the DL website (www.biomerieux-usa.com/diversilab) was used.
The distributions of the 986 S. aureus isolates are shown in Table 1. All isolates were obtained in general hospitals (400 isolates, 40.6%), hospitals and clinics (475 isolates, 48.2%), or geriatric hospitals (111 isolates, 11.3%). Of these, 803 MRSA isolates (81.4% of the total isolates) were identified through oxacillin screening and with the mecA PCR method. Among all MRSA infections, 294 (36.6%) isolates were identified as CA-MRSAs with non-multidrug resistance (non-MDR; defined as susceptible to ≥2 tested non-β-lactam antibiotic drugs), and the rest (509 [63.4%]) were classified as HA-MRSAs. Most of the identified HA-MRSAs were recovered from invasive infections (e.g., pus [n=150], wounds [n=129], sputum [n=97], blood [n=50], urine [n=36], ear fluid [n=14], pleural fluid [n=12]), although 47 isolates were recovered from nasal swabs (data not shown).
On MLST analysis, SCCmec types for all MRSAs were characterized by four major clones and the results are shown in Fig. 1 and 2: CC5 belonging to the SCCmec II type (including IIA and IIvariant), CC8 belonging to the SCCmec III type (including IIIA and IIIvariant), and CC72 and CC1 belonging to the SCCmec IV type (including IVA). All MRSAs were clonally distributed throughout the country and hospitals. Regionally, five clones-CC1, CC5, CC8, CC72, and CC89-made up the majority of the isolates, whereas CC30, CC59, CC121, and CC608 were less frequent. In general hospitals, CC5 and CC8 were distributed primarily in Seoul. CC72 was the main clone distributed in hospitals and clinics at Chungcheongnam-do/Daejeon (CN/DJ), and CC5 was widespread in geriatric hospitals at Jeollanam-do (JN) and Gyeongsangnam-do (GN). The presence of lukF-PV and lukS-PV genes encoding the components of the PVL toxin was detected in all isolates. CA-MRSA isolates most commonly had SCCmec II and IV type traits and no positive PVL genes (data not shown).
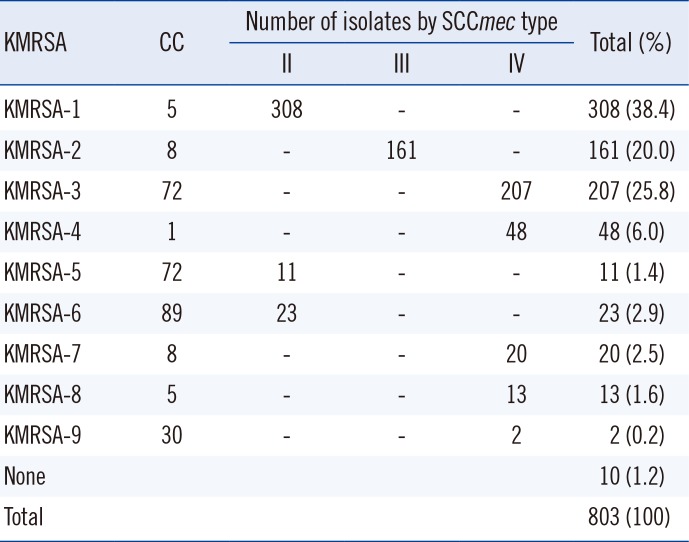
The clones and mec types were classified as KMRSA-1 through KMRSA-9 and summarized in Table 2. KMRSA-1 was the most frequent, identified in 308 isolates (38.4% of all MRSAs), and has the genetic characteristics of CC5 and the SCCmec II type. In contrast, 13 isolates (1.6%) had the genetic characteristics of CC5 and the SCCmec IV type; therefore, these were identified as KMRSA-8. A total of 161 isolates (20.0%) had the genetic characteristics of CC8 and the SCCmec III type, and thus were identified as KMRSA-2, whereas 20 isolates with the SCCmec IV type were designated KMRSA-7. KMRSA-3 was identified in 207 isolates (25.8%) with the genetic characteristics of CC72 and the SCCmec IV type, whereas 11 isolates (1.4%) with the SCCmec II type were designated KMRSA-5. KMRSA-4 (CC1 and the SCCmec IV type) comprised 48 isolates (6.0%). KMRSA-6 was identified in 23 isolates (2.9%) with the genetic characteristics of CC89 and the SCCmec II type, and KMRSA-9 was identified in two isolates (0.2%) with the genetic characteristics of CC30 and the SCCmec IV type.
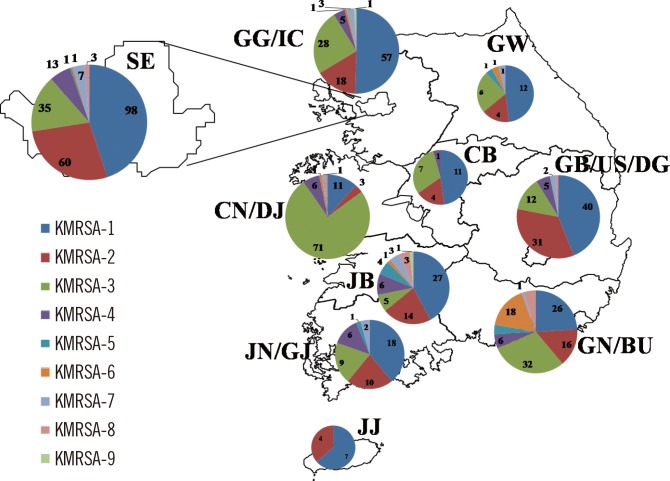
KMRSA was distributed in 10 regions across the country in the following numbers: Seoul, n=222; Gyeonggi-do/Incheon, n=113; Gangwon-do, n=25; Chungcheongbuk-do, n=23; Chungcheongnam-do/Daejeon, n=94; Jeollabuk-do, n=68; Jeollanam-do/Gwangju, n=47; Gyeongsangbuk-do/Ulsan/Daegu, n=91; Gyeongsangnam-do/Busan, n=109; and Jejudo, n=11 (Fig. 3). KMRSA-1, KMRSA-2, and KMRSA-3 were prevalent throughout most of the country, including Seoul and Gyeonggi-do Province. KMRSA-3, KMRSA-4, and KMRSA-6 had higher distribution ratios in Daejeon, Jeollanam-do and Gwangju, and Gyeongsangnam-do, respectively.
We analyzed the DL patterns of KMRSA strains by using the DL MRSA library STAT sheet, which comprises 70 samples with representative USA pulsed-field gel electrophoretic (PFGE) and SCCmec types. A DL typing dendrogram representing nine KMRSA strains is shown in Fig. 4. The average similarity among KMRSA-1 through KMRSA-8 was more than 84%; KMRSA-9 differed from the other strains.
In this study, we investigated the molecular and genetic characteristics and trends of national staphylococcal clones in hospital settings in Korea over an 8-yr period. We isolated an average of 124 strains per year nationally, and despite the limitation of the small sample numbers, we evaluated the clinical importance of the isolates.
The distribution ratio of the collected MRSA isolates (up to 81.4%) was relatively higher than that of foreign MRSA [13]. In particular, the average MRSA distribution for all medical settings, including hospitals, was more than 70% for all hospitals and more than 80% for LTCFs (data not shown). These results are consistent with the resistance rates reported by the Korean Antimicrobial Resistance Monitoring System (KARMS) and the Korean Nationwide Surveillance of Antimicrobial Resistance (KONSAR) [14]. In clinics, MRSA rates were also relatively higher (60%) than the average of 40% reported by KARMS.
In this study, we categorized KMRSA strains according to sequence type (ST), SCCmec type, and the presence of PVL. From this classification, we categorized the strains of MRSA on the basis of the genetic characteristics and compared domestic MRSA with MRSA strains that are epidemic in the USA, United Kingdom (UK), and Canada. Frequently observed MRSA strains, including USA100, USA400, and eclone-1, have been identified in the USA and Europe. Of the epidemic MRSA strains designated in this study, KMRSA-1, which had the highest distribution ratio, showed genetic characteristics identical to those of USA100 and Canada MRSA-2 (ST5 and SCCmec type II). This clone is distributed worldwide, including in Korea. KMRSA-8, which has genetic similarity to KMRSA-1, was less frequently identified but was confirmed in some areas overseas.
KMRSA-3 was identical to USA700 and was derived from local communities. Moreover, this clone had genetic similarity to the USA300 clone of PVL-positive MRSA. However, only three isolates of this domestic clone were identified, which suggested that it is not a common type. KMRSA-2 (CC8 and the SCCmec III type) was the third epidemic clone and differed from epidemic clones of the SCCmec IV type found overseas. KMRSA-7, which was genotypically identical to KMRSA-2, was uncommonly distributed and was the same as USA500 and UK-CC8. KMRSA-4 was identical to clones USA400 and UK-CC1, and KMRSA-9 was identical to USA200 and UK-CC30. KMRSA-4 and KMRSA-9 were also separated by the individual clonal types CC1 and CC30 in the community and hospitals.
KMRSA-1, KMRSA-2, and KMRSA-3 had the widest distribution nationwide and were distributed evenly in several hospitals in Gyeongsangnam-do and Busan. In particular, KMRSA-3 had relatively wide distribution in Chungcheongnam-do and Daejeon. This result may be attributable to the concentration of KMRSA-3 in specific pediatric hospitals in Daejeon, but the possibility that KMRSA-3 was trending in Daejeon at the time of the study also cannot be ruled out.
The most common lineages in the USA are CC1 (USA400) and CC8 (USA300), which usually carry SCCmec type IV and PVL-encoding genes and are becoming the subject of intense research [315]. The four PVL-positive MRSA stains in this study had ST30 (one isolate) or ST8 (three isolates) characteristics. Of these, three isolates showed the molecular characteristics of an HA-MRSA of SCCmec III type with the staphylococcal enterotoxin genes sea and seb, and the other showed molecular characteristics of a CA-MRSA of SCCmec IV type with the staphylococcal enterotoxin genes seg and sei. These results were confirmed by the observation that the USA clone of a PVL-positive community epidemic was also identified in the Korean hospital environment during various periods. Thus, these strains exhibited some genetic differences from strains in dominant countries, including the USA, Canada, and Latin America [16].
In summary, the methicillin resistance rate of staphylococcal isolates in Korea was slightly higher than that in other countries. Comparisons of domestic and foreign MRSA clones showed similarities in three types, and the genetic similarities to foreign clones were characterized. Some of these strains were related to those that had spread overseas, whereas others (e.g., KMRSA-2) had genetic traits, such as different SCCmec structures, that were nationally distributed. In addition, we confirmed that PVL-positive MRSA strains were not HA strains distributed in hospitals in Korea. Therefore, we believe that this comparative study of the genetic characteristics of epidemic MRSA clones will be a useful resource for the establishment of infection control measures in future multidrug-resistant staphylococcal outbreaks and in cases of newly occurring resistant strains related to the influx of foreign strains. Our results have also identified additional target strains that may be helpful in future monitoring efforts.
Acknowledgments
This study was supported by a grant from the Korea Centers for Disease Control and Prevention (2012-N44001-00).
References
1. Korea Centers for Disease Control and Prevention KCDC. Korean antimicrobial resistance monitoring system annual report. Cheongju, Korea: The Centers;2011.
2. Larsen AR, Stegger M, Böcher S, Sørum M, Monnet DL, Skov RL. Emergence and characterization of community-associated methicillin-resistant Staphylococcus aureus infections in Denmark, 1999 to 2006. J Clin Microbiol. 2009; 47:73–78. PMID: 18971362.
3. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003; 41:5113–5120. PMID: 14605147.
4. Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011; 66(S4):iv43–iv48. PMID: 21521706.
5. Kramer A, Wagenvoort H, Åhrén C, Daniels-Haardt I, Hartemann P, Kobayashi H, et al. Epidemiology of MRSA and current strategies in Europe and Japan. GMS Krankenhhyg Interdiszip. 2010; 5:Doc01. PMID: 20204100.
6. Centers for Diseases Control and Prevention. Antibiotic resistance threats in the United States, 2013. Updated on Jul 2014. http://www.cdc.gov/drugresistance/threat-report-2013/.
7. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement, M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute;2014.
8. Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002; 46:2155–2161. PMID: 12069968.
9. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999; 29:1128–1132. PMID: 10524952.
10. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–1015. PMID: 10698988.
11. Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J Clin Microbiol. 2005; 43:1187–1192. PMID: 15750081.
12. Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE Jr. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J Clin Microbiol. 2009; 47:2452–2457. PMID: 19553588.
13. Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010; 15:19688. PMID: 20961515.
14. Lee Y, Kim YA, Song W, Lee H, Lee HS, Jang SJ, et al. Recent trends in antimicrobial resistance in intensive care units in Korea. Korean J Nosocomial Infect Control. 2014; 19:29–36.
15. Jones MB, Montgomery CP, Boyle-Vavra S, Shatzkes K, Maybank R, Frank BC, et al. Genomic and transcriptomic differences in community acquired methicillin resistant Staphylococcus aureus USA300 and USA400 strains. BMC Genomics. 2014; 15:1145. PMID: 25527145.
16. Nimmo GR. USA300 abroad: global spread of a virulent strain of community associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012; 18:725–734. PMID: 22448902.
Fig. 1
Distribution of clonal complexes (CCs) by staphylococcal cassette chromosome mec (SCCmec) type. Singleton refers to sequence types (STs) that could not be assigned to any group.
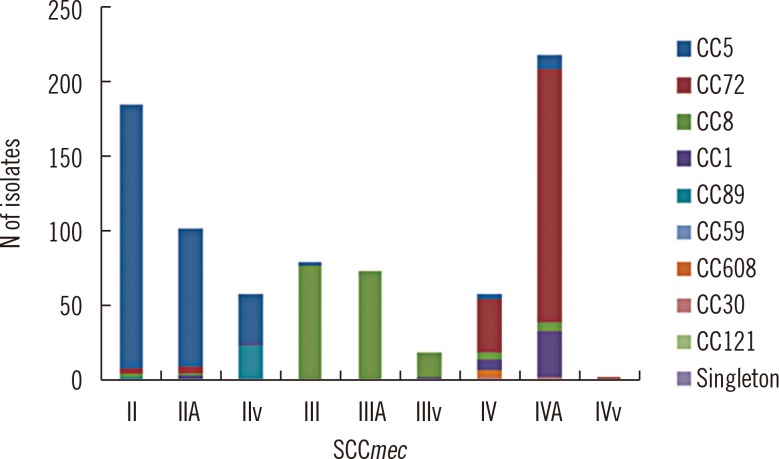
Fig. 2
Clonal distribution of all methicillin-resistant Staphylococcus aureus (MRSA) isolates by district. The prevalence of CCs in (A) general hospitals, (B) hospitals and clinics, and (C) geriatric hospitals is shown. Singleton refers to STs that could not be assigned to any group.
Abbreviations: CB, Chungcheongbuk-do; CN/DJ, Chungcheongnam-do/Daejeon; GB/US/DG, Gyeongsangbuk-do/Ulsan/Daegu; GG/IC, Gyeonggi-do/Incheon; GN/BU, Gyeongsangnam-do/Busan; GW, Gangwon-do; JB, Jeollabuk-do; JJ, Jejudo; JN/GJ, Jeollanam-do/Gwangju; SE, Seoul.
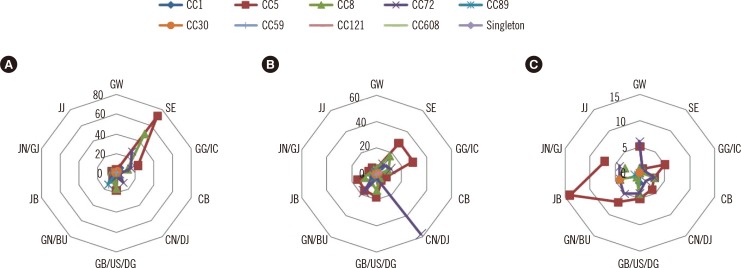
Fig. 4
Dendrogram analysis of nine representative Korean MRSA (KMRSA) strains (circled) obtained by using the DiversiLab (DL; bioMérieux, Mercy l'Etoile, France) MRSA library sheets. The gel-like image generated by the DL software illustrates band similarities.
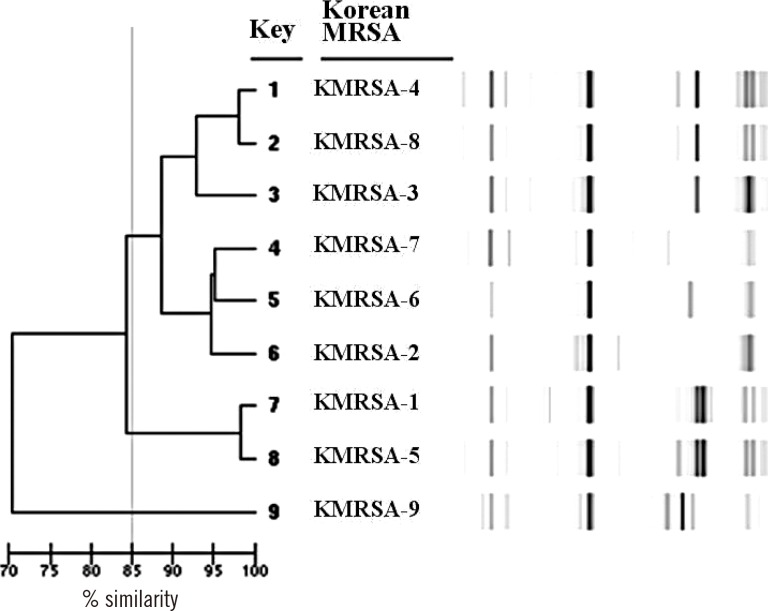
Table 1
Number of Staphylococcus aureus isolates collected in the study
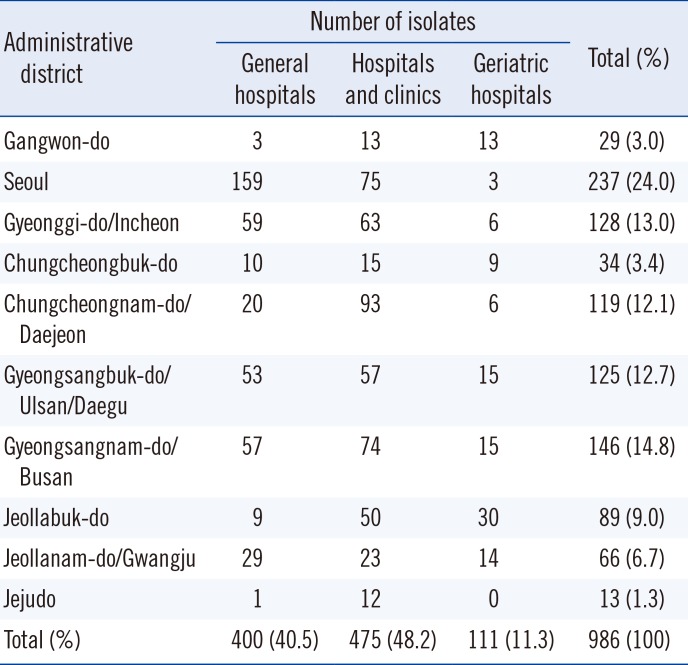
Table 2
Classification of KMRSA




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download