Abstract
Background
Methods
Results
Conclusions
Acknowledgments
References
Fig. 1
Frequency distribution of TP53 germline mutations by codon. (A) Data reproduced from the International Agency for Research on Cancer (IARC) TP53 database (version R18, April 2016, http://p53.iarc.fr/) [7]. (B) Data from Korean patients with Li-Fraumeni syndrome. Figures above the vertical line represent the codon locations of the mutations.
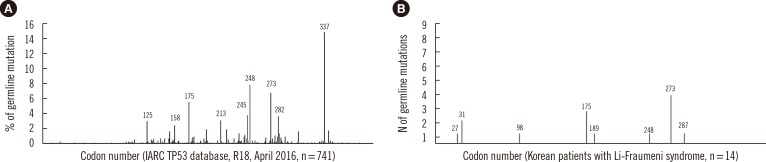
Fig. 2
Sequence chromatogram of two novel frameshift mutations in TP53. (A) Heterozygous mutation of c.293delC (p.Pro98Leufs*25) identified in patient 6, who was treated for adrenocortical carcinoma, osteosarcoma, brain cancer, and therapy-related neoplasm. (B) Heterozygous mutation of c.78delT (p.Pro27Leufs*17) identified in patient 7 with multiple primary tumors comprising left breast cancer (28 yr), neurofibroma (33 yr), right breast cancer (38 yr), right nasal cavity squamous cell carcinoma (38 yr), and lung adenocarcinoma (43 yr). Forward and reverse sequences are shown at the top and bottom, respectively.
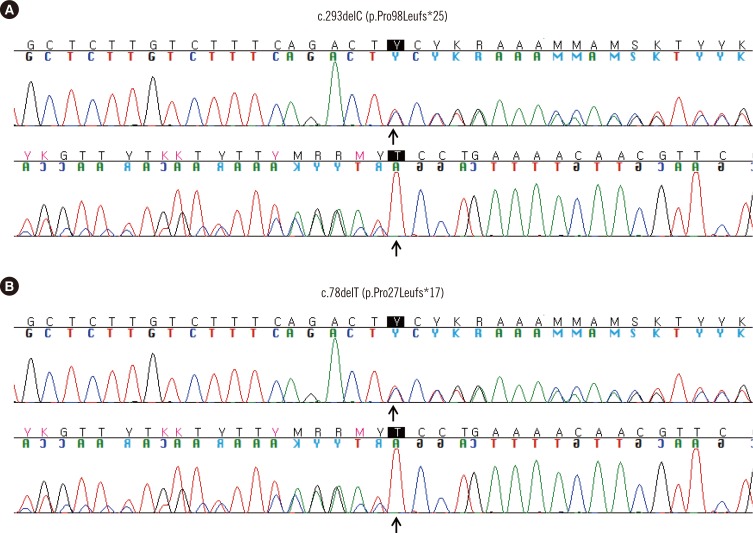
Table 1
Clinical and TP53 mutation data of the 14 Korean patients with Li-Fraumeni syndrome
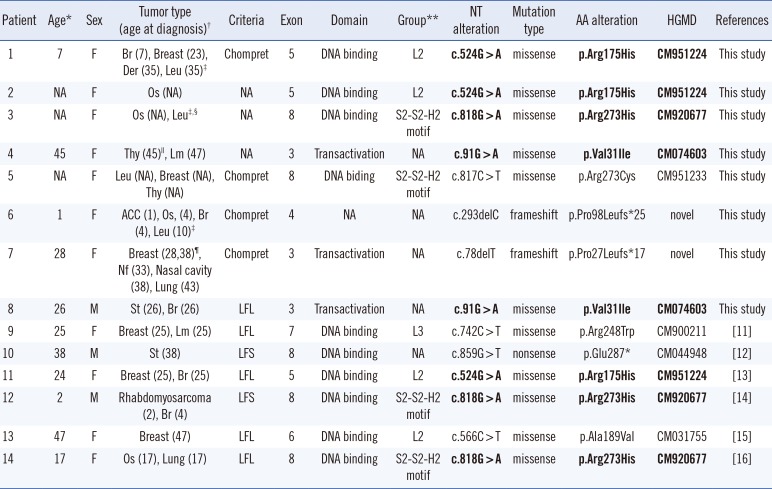
| Patient | Age* | Sex | Tumor type (age at diagnosis)† | Criteria | Exon | Domain | Group** | NT alteration | Mutation type | AA alteration | HGMD | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | F | Br (7), Breast (23), Der (35), Leu (35)‡ | Chompret | 5 | DNA binding | L2 | c.524G > A | missense | p.Arg175His | CM951224 | This study |
| 2 | NA | F | Os (NA) | NA | 5 | DNA binding | L2 | c.524G > A | missense | p.Arg175His | CM951224 | This study |
| 3 | NA | F | Os (NA), Leu‡,§ | NA | 8 | DNA binding | S2-S2-H2 motif | c.818G > A | missense | p.Arg273His | CM920677 | This study |
| 4 | 45 | F | Thy (45)∥, Lm (47) | NA | 3 | Transactivation | NA | c.91G > A | missense | p.Val31Ile | CM074603 | This study |
| 5 | NA | F | Leu (NA), Breast (NA), Thy (NA) | Chompret | 8 | DNA biding | S2-S2-H2 motif | c.817C > T | missense | p.Arg273Cys | CM951233 | This study |
| 6 | 1 | F | ACC (1), Os, (4), Br (4), Leu (10)‡ | Chompret | 4 | NA | NA | c.293delC | frameshift | p.Pro98Leufs*25 | novel | This study |
| 7 | 28 | F | Breast (28,38)¶, Nf (33), Nasal cavity (38), Lung (43) | Chompret | 3 | Transactivation | NA | c.78delT | frameshift | p.Pro27Leufs*17 | novel | This study |
| 8 | 26 | M | St (26), Br (26) | LFL | 3 | Transactivation | NA | c.91G > A | missense | p.Val31Ile | CM074603 | This study |
| 9 | 25 | F | Breast (25), Lm (25) | LFL | 7 | DNA binding | L3 | c.742C > T | missense | p.Arg248Trp | CM900211 | [11] |
| 10 | 38 | M | St (38) | LFS | 8 | DNA binding | NA | c.859G > T | nonsense | p.Glu287* | CM044948 | [12] |
| 11 | 24 | F | Breast (25), Br (25) | LFL | 5 | DNA binding | L2 | c.524G > A | missense | p.Arg175His | CM951224 | [13] |
| 12 | 2 | M | Rhabdomyosarcoma (2), Br (4) | LFS | 8 | DNA binding | S2-S2-H2 motif | c.818G > A | missense | p.Arg273His | CM920677 | [14] |
| 13 | 47 | F | Breast (47) | LFL | 6 | DNA binding | L2 | c.566C > T | missense | p.Ala189Val | CM031755 | [15] |
| 14 | 17 | F | Os (17), Lung (17) | LFL | 8 | DNA binding | S2-S2-H2 motif | c.818G > A | missense | p.Arg273His | CM920677 | [16] |
The reference sequence of TP53 was NM_000546.4. Recurrent mutations are in bold. All mutations were heterozygous.
*Age at first tumor diagnosis; †In the order of tumor diagnosis; ‡Therapy-related myeloid neoplasm; §positive for chromosomal breakage test; ∥BRAF V600E mutation positive case; ¶BRCA 1 and BRCA 2 mutation negative and positive for hormone receptors (estrogen and progesterone) and ERBB2 (formerly HER2/neu); **Based on structural data from [817].
Abbreviations: NT, nucleotide; AA, Amino acid; LFS, Li-Fraumeni syndrome; LFL, Li-Fraumeni syndrome like; NA, not applicable, Br; brain; Der, dermatofibrosarcoma; Leu, leukemia; Os, Osteosarcoma; Thy, thyroid carcinoma; Lm, leiomyoma; ACC, adrenocortical carcinoma; Nf, neurofibroma; St, stomach; HGMD, Human Genome Mutation Database.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download