INTRODUCTION
During the 2015 outbreak of Middle East Respiratory Syndrome coronavirus (MERS-CoV), a total of six different commercial MERS-CoV RNA detection kits based on real-time reverse-transcription polymerase chain reaction (rRT-PCR) were available in Korea. Of those, the PowerChek MERS Real-time PCR (Kogene Biotech, Seoul, Korea), DiaPlexQ MERS Virus Detection (SolGent, Daejeon, Korea), and Anyplex II MERS-CoV (
upE) Real-time Detection kits (Seegene, Seoul, Korea) comprised two steps: single gene targeting of the region upstream of the envelope gene (
upE) for screening, and multiple gene targeting of both
upE and open reading frame 1a (
ORF1a) for confirmation. The AccuPower MERS-CoV Real Time RT-PCR (Bioneer, Daejeon, Korea), LightMix Molecular Dx MERS-CoV
upE/ORF1a (Roche Molecular Diagnostics, Basel, Switzerland), and UltraFast LabChip MERS-CoV Real-time PCR kits (Nanobiosys, Seoul, Korea) used one step to simultaneously detect both
upE and
ORF1a using two single gene-targeting reagents. None of these kits have been approved for diagnostic use; however, they were urgently introduced into clinical laboratories on June 4, 2015 because the timely diagnosis of MERS-CoV infections was essential during the nationwide MERS-CoV outbreak in Korea [
12].
The WHO and United States Centers for Disease Control and Prevention (US CDC) provided guidelines for the molecular diagnosis of MERS-CoV [
34], and since June 6, 2013 the US CDC has made novel coronavirus rRT-PCR assays [
5] available free of charge under emergency use authorization [
6]. Although at least three commercial rRT-PCR assays for MERS-CoV detection were available from Altona Diagnostics, Fast Track Diagnostics [
3], and PrimerDesign (
http://www.genesig.com) before the 2015 outbreak in Korea, only RealStar MERS-CoV (Altona Diagnostics, Hamburg, Germany) had been approved for the
in vitro diagnosis of MERS-CoV by Conformité Européenne (CE) and authorized for emergency use only in the United States. Therefore, all six commercial kits evaluated in this study had not been validated for diagnostic use. This study was designed to analytically and clinically validate the six above-mentioned commercial MERS CoV RNA detection kits.
METHODS
During July 6-10, 2015, each kit was validated by using the equipment recommended by each manufacturer (
Table 1). To determine analytical sensitivity, the limits of detection (LOD) with 95% probability values was determined by using
upE and
ORF1a RNA transcripts supplied by the Institute of Virology, University of Bonn Medical Centre [
7]. The original concentration of both RNA transcripts was 1.0×10
5 copies/µL. These were diluted to six concentrations in 0.5-log steps from 100 to 0.3 copies/reaction, and kits were tested by using 5-8-µL samples of RNA eluates per reaction. For the Nanobiosys kit, which used 2.4-µL samples per reaction, a 0.5-log higher concentration was added for the LOD validation. Each concentration was tested by using 16 replicates, with the exception of PowerChek, for which 12 replicates were used. A probit regression analysis in R Studio (R Studio Inc.;
https://www.rstudio.com/) was performed to determine the 95% cut-off values. The PowerChek, AccuPower, LightMix, and UltraFast LabChip kits used the primers and probes from the WHO-recommended rRT-PCR assay [
78]. The primers and probes used by the DiaPlexQ and Anyplex kits were modified from the WHO-recommended rRT-PCR assay, but covered almost the same regions of
upE and
ORF1a (personal communication with confidentiality of the sequences). However, the Anyplex kit was validated only for
upE because the oligonucleotide-binding site for
ORF1a was beyond the span of the RNA transcripts used in this study.
To evaluate the analytical and clinical specificity of the kits, 28 respiratory virus-positive nasopharyngeal swabs were used to determine cross-reactions with human RNA or other respiratory viruses, including human coronaviruses. Using the Anyplex II RV16 kit (Seegene) with duplicate specimen preparations, these specimens were confirmed as positive for only single species of the following viruses: influenza virus A (n=2), influenza virus B (n=2), human parainfluenza virus 1 (n=2), human parainfluenza virus 2 (n=2), human parainfluenza virus 3 (n=2), respiratory syncytial virus A (n=2), respiratory syncytial virus B (n=2), human adenovirus (n=2), human bocavirus (n=2), human metapneumovirus (n=2), human rhinovirus (n=2), human coronavirus 229E (n=2), human coronavirus OC43 (n=2), and human coronavirus NL63 (n=2).
To evaluate the clinical sensitivity of the assays, 18 specimens from the lower respiratory tract, including 14 known positive specimens, were obtained from five institutions. The RNA eluates of nine specimens that remained after initial clinical MERS-CoV rRT-PCR testing were used following storage of <2 months at -70℃ at each institution. The other nine specimens were manually prepared in our laboratory without any pretreatment and were found to be inadequate for evaluating clinical sensitivity owing to the extremely high levels of PCR inhibition. Therefore, only the first nine specimens were used to evaluate clinical sensitivity, and the other nine specimens with high inhibition were used to evaluate the effects of PCR inhibition on positive results. In addition to determining the numbers of positives and negatives, the cycle threshold (Ct) values of the target genes and internal control were analyzed.
RESULTS
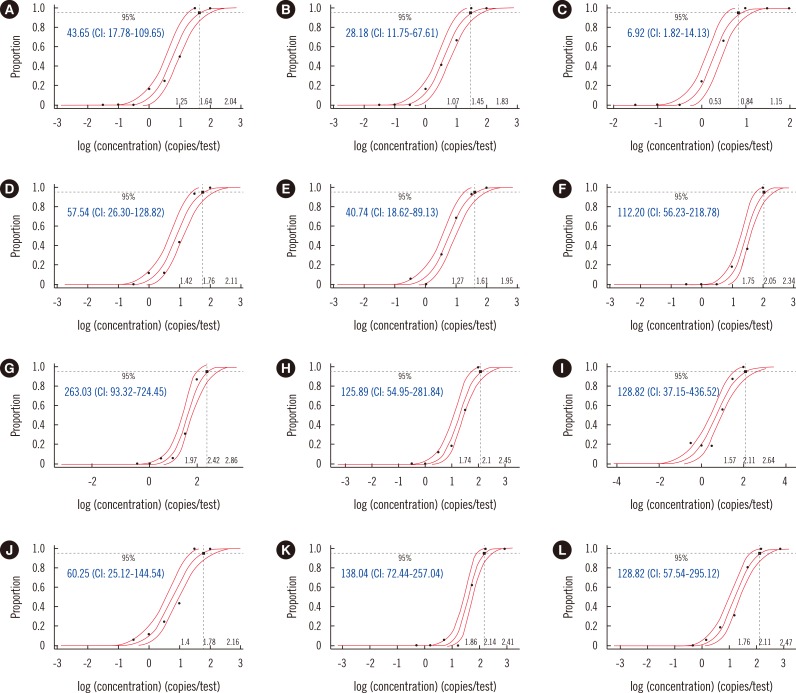
The LODs for
upE varied from 21.88 to 263.03 copies/reaction, and those of
ORF1a varied from 6.92 to 128.82 copies/reaction (
Fig. 1). According to the probit regression analysis, the 95% confidence intervals (CI) for
upE and
ORF1a were found to overlap among the tested kits, with the exception of
ORF1a by the PowerChek kit (
Fig. 2). The LODs for
upE using both the single and multiple gene-targeting formats of the 2-step kits were 1.64 and 1.45 log copies/reaction for the PowerChek kit and 1.76 and 1.61 log copies/reaction for the DiaPlexQ kit (
Fig. 2). The LOD for
upE was 1.34 log copies/reaction for the Anyplex kit, but no CI value could be calculated because there were no positive reactions at 1.0 log copy/reaction, while all 16 replicate specimens were positive at 1.5 log copies/reaction. In contrast, although the LODs for
upE and
ORF1a using the LightMix kit were 2.11 log copies/reaction and 1.78 log copies/reaction, respectively, trailing of positives was observed at much lower concentrations than these LODs (
Fig. 1). The LODs for
upE of three different 1-step kits, the AccuPower, Light Mix, and Ultrafast kits, were >2.0 log copies/reaction, which was less sensitive than those of three different 2-step kits; however, this difference was not significant (
Fig. 2). The LODs for
ORF1a varied less—i.e., from 1.78 to 2.11 log copies/reaction—with the exception of the PowerChek kit, which was considerably lower at 0.84 log copies/reaction (
Fig. 2). None of the kits tested in this study showed cross-reactivity with other respiratory viruses.
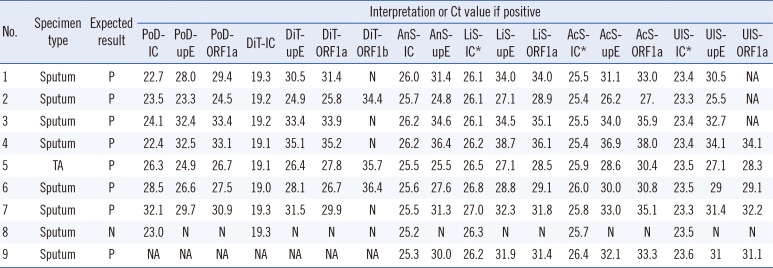
Based on validation tests including 28 specificity panel and nine clinical specimens, the clinical sensitivities of detecting
upE and
ORF1a were 100% (95% CI, 0.60-1.00) for all study kits. Specificity was 100% (95% CI, 0.79-1.00). The positive samples showed a wide range of Ct values, with values between 7.6 and 11.6 resulting from all kits (
Table 2). The Ct values of the internal control varied most with PowerChek, from 22.7-32.1, while those of DiaPlexQ, Anyplex, AccuPower, LightMix, and UltraFast all varied by <1.0 (
Table 2).
ORF1b, tested by the DiaPlexQ kit, was detected in only five positive specimens (62.5%). The
upE results of the UltraFast kit were not available for three positive specimens because one of the capillary PCR chambers holding these specimens leaked.
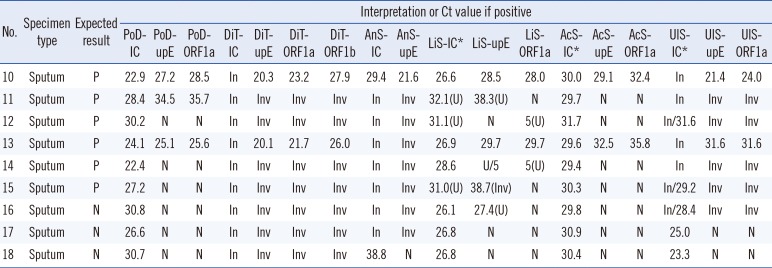
The effect of PCR inhibition determined by testing the nine high inhibition specimens revealed that 3-5 of the six known positive specimens were found negative or invalid by each kit (
Table 3). The Ct values of the internal controls from all specimens were high or extended beyond the total amplification cycles, and, therefore, false negative results were eliminated owing to internal control failure, except for those of the PowerChek and AccuPower kits. With the AccuPower kit, the mean internal control Ct value for the high inhibition specimens was considerably prolonged to 30.1±0.69 compared with 25.7±0.31 for the sensitivity test specimens; however, there was no established cut-off point, indicating that PCR inhibition was not acceptable in the PCR reaction. On the other hand, Ct values for the sensitivity test specimens and the high inhibition specimens using the PowerChek kit were not significantly different at 25.3±3.2 and 27.0±3.1, respectively. The LightMix kit produced two uncertain results with known positive specimens and one uncertain result with a known negative specimen (
Table 3).
There were no positive results detected by any of the kits among the four negative specimens included in the sensitivity test and high inhibition panels (
Tables 2 and
3).
DISCUSSION
The LOD for the
upE and
ORF1a rRT-PCR kits was analyzed with the same RNA transcripts used to evaluate RealStar MERS-CoV (Altona Diagnostics), which is the only kit thus far approved for diagnosis [
9]. When comparing the analytical sensitivities of assay systems, it is important to use consistent evaluation conditions; moreover, the quality of the source material is critical. It is often difficult to find traceable source materials for the molecular diagnosis of viral infections, as only a few international standards have been established to date [
101112]. As with RealStar MERS-CoV [
9], all target gene-binding sites for
upE,
ORF1a, and
ORF1b used in the study kits were based on the oligonucleotide sequences of WHO-recommended primers and probes [
3], and the
upE and
ORF1a target sites were located within the RNA transcripts used in this study. Whole genome sequences of MERS-CoV isolates obtained during the 2015 Korean outbreak were determined by the Korea National Institute of Health and Seoul National University (gb|KT029139.1|) and the Guangdong Provincial Center for Disease Control and Prevention (gb|KT036372.1). Both sequences closely clustered with a strain isolated during the spring 2015 outbreak in Riyadh, Saudi Arabia (gb|KT026454.1|) [
13]. Therefore, the primers and probes used in this study were most likely not affected by sequence variations in the MERS-CoV strains circulating in this outbreak. The RNA transcripts used to assess the analytical sensitivity in detecting
upE and
ORF1a [
9] were not expected to have target mismatches with the primers and probes used in this study. Therefore, the LOD of all kits was analyzed by using identical source materials, and the data were comparable among various kits as well as with data from previous evaluations of RealStar MERS-CoV [
9].
The LODs for
upE of the kits assessed in this study indicated approximately 10-fold lower sensitivity than the previously reported 95% cut-off value of 5.3 copies/reaction for RealStar MERS-CoV [
9]. The overall sensitivities for detecting
upE of the kits evaluated herein were consistently lower than that of the RealStar MERS-CoV kit (
Fig. 2). Only the LOD for
ORF1a using the PowerChek duplex kit was similar to that of the RealStar kit: 9.3 copies/reaction using the RealStar MERS-CoV kit in comparison with 6.9 copies/reaction using the PowerChek kit. Therefore, the lower sensitivities of the kits evaluated in comparison with that of RealStar MERS-CoV could be attributed to their performances rather than instability in source materials. In comparison with RealStar MERS-CoV [
9], all of the kits analyzed herein have room for improvement in their sensitivity, with the exception of
ORF1a testing with the PowerChek kit. Unlike the other tested kits, the LightMix kit demonstrated a markedly trailing tendency to yield positive results down to much lower concentrations than its LOD and an uncertain result to even a known-negative specimen (
Fig. 1). These findings suggest that the effort to increase sensitivity may result in more uncertainty and therefore negatively impact specificity. The previous study on the clinical sensitivity of RealStar MERS-CoV produced four discrepant results among 19 specimens, of which three specimens (oral, nasal, and urine specimens) showed high Ct values using the RealStar kit, but not using the comparison assay (and vice versa for one nasal specimen) [
9]. This could be a result of RealStar MERS-CoV having a higher sensitivity or lower specificity than the other assay. Using the 2-step kits tested in this study, the LODs for
upE using multiple gene-targeting formats were comparable to those obtained using single gene-targeting formats (
Fig. 2). Therefore, implementing a 2-step approach, including performing the first screening step with a single gene-targeting format, was not beneficial in terms of sensitivity.
In the present study, the LOD was analyzed by using a 0.5-log dilution series because a 0.5-log dilution is considered "within physiological variation or not significant" in viral kinetics [
14]. However, this was limited in that the 0.5-log dilution scale was too wide to estimate the CI value for the Anyplex kit (
Fig. 1), and estimation of the LOD by probit regression analysis seemed to be less appropriate for the LightMix kit with a trailing tendency to much lower concentrations. The LOD for
ORF1b using the DiaPlexQ kit was not measured, but the low sensitivity of this kit in detecting
ORF1b in clinical specimens is consistent with a previous report, in which testing for
ORF1b was less sensitive than testing for
ORF1a or
upE [
78].
No cross-reactivity was observed with other respiratory viruses, including human coronavirus, in any assays included in this study. These findings are consistent with previous evaluations that used the primers and probes recommended by WHO [
789]. Therefore, all assay systems demonstrated adequate specificity.
The clinical sensitivities of the kits were all 100% (
Table 2). Because the sensitivity test panel comprised various positive specimens with a wide range of Ct values, this high sensitivity validated their diagnostic performance for MERS-CoV infection, particularly from sputum specimens, although the number of positive specimens was small. One of the strengths in the present study was determining the effects of PCR inhibition on clinical sensitivity and the importance of internal controls (
Table 3). The internal controls of the PowerChek kit were not affected by the inhibition. This could be because the PowerChek kit uses a less sensitive internal control that is intrinsic to human cells, although the PCR efficiency of this kit itself was also less affected. The positivity rates for kits when testing the high inhibition panel correlated with their analytical sensitivities, as only PowerChek detected 50% of known-positive specimens. Based on these findings, it is clear that clinical sensitivity can be significantly affected by the presence of PCR inhibitors, and analytical sensitivity may be affected by PCR inhibition-prone specimens such as sputum specimens. Therefore, optimization of the pretreatment and RNA extraction procedures is necessary to improve the sensitivity of MERS-CoV molecular diagnostics, especially for testing sputum specimens.
There were some noteworthy limitations of this study, including: 1) the number of specimens used to test clinical sensitivity was relatively small; 2) the reproducibility of the assay results was not validated; and 3) the effects of inhibition were qualitatively estimated. In conclusion, the commercial kits evaluated in this study demonstrated variable analytical sensitivities; however, the overall clinical sensitivity and specificity were same and were sufficient for diagnosing MERS-CoV infection. The performance of the individual kits could be improved in terms of their analytical sensitivity, and identification of PCR inhibition with the use of appropriate internal controls is essential.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download