Dear Editor,
KIT (also known as c-KIT) is a receptor tyrosine kinase expressed on various cell types [1]. KIT mutations are detected in approximately 5% of patients newly diagnosed with acute myeloid leukemia (AML) and in 20-30% of patients with core-binding factor AML [23]. KIT mutations are predominantly presented as one or two amino acid substitutions affecting codon 816. The most frequent mutation type, D816V, is detected in about 40% of cases with AML, where KIT is mutated [4]. In AML with t(8;21), the presence of KIT mutation at codon 816 is associated with a high white blood cell (WBC) count at diagnosis, a high incidence of extramedullary leukemia, and high risk of relapse during the course of the disease [5]. Other less frequent types of KIT mutation are D816Y, D816H, and D816I. KIT mutations present a prognostic significance. The National Comprehensive Cancer Network categorized AML with either t(8;21) or inv(16)/t(16;16) combined with KIT mutations as the intermediate-risk group [6]. We report a novel INDEL mutation in KIT codon 816, discovered in a woman with AML with t(8;21)(q22;q22); RUNX1-RUNX1T1.
A 35-yr-old woman was admitted for purpura in both lower limbs. The initial complete blood cell count indicated: WBC count, 20.1×109/L; hemoglobin, 8.2 g/dL; and platelet count, 15×109/L. A peripheral blood smear revealed leukocytosis with 45% of blasts. The bone marrow (BM) aspirate smear and cytochemical stains showed an increased number of myeloblasts (68%) and other myeloid precursors (Fig. 1A). The 197 bp-sized transcript of RUNX1 exon 6/RUNX1T1 exon 2 was detected in a multiplex PCR analysis of BM aspirates. In the cytogenetic study, the karyotype was 46,XX,t(8;21)(q22;q22)[18]/46,XX[2]. The patient was diagnosed with AML with t(8;21)(q22;q22); RUNX1-RUNX1T1. In the screening test for KIT mutations using a real-time allele-specific PCR kit (Real-Q C-KIT; BioSewoom, Seoul, Korea), a mutation was detected, which was not the D816V, D816Y, or D816H type (Fig. 1B). Conventional sequencing was performed to identify the mutation. Unlike the common mutation types presenting one or two ambiguous bases in codon 816, the sequencing result showed five consecutive ambiguous bases through codon 815–817 (Fig. 1C). Allele separation by T-vector cloning using pGEM-T Easy Vector Systems (Promega, Madison, WI, USA) and additional conventional sequencing were performed and a missense INDEL mutation NM_000222.2:c.2445_2449delAGACAinsGTTAC (p.D816_I817delinsLL) was confirmed. To the best of our knowledge, this mutation has not yet been reported.
Three in-silico prediction tools were utilized to predict the effect of the mutation on the KIT protein function and stability (Table 1). Using SIFT (Sorting Tolerant From Intolerant, version 1.03, http://sift.jcvi.org/; J. Craig Venter Institute, La Jolla, CA, USA) [7], exploiting the degree of conservation of amino acid residues derived from functionally related protein sequences, D816L, I817L, and D816V mutations were predicted to be 'damaging', with a SIFT score equal to 0 (the amino acid substitution is predicted as 'damaging' if the score is ≤0.05). Using PolyPhen-2 (Polymorphism Phenotyping v2, version 2.2.2, http://genetics.bwh.harvard.edu/pph2/; Division of Genetics, Brigham & Women's Hospital, Harvard Medical School, Boston, MA, USA) [8] to predict the possible impact of an amino acid substitution on the structure and function of a protein using several features, including sequences, phylogenetic, and structural information, D816L and I817L were predicted to be 'probably damaging' with a score of 1.000, and 'possibly damaging' with a score of 0.955, respectively. By comparison, the D816V mutation was predicted to be 'probably damaging', with a score of 0.999. Using SABLE (Accurate prediction of Solvent AccessiBiLitiEs, version 2, http://sable.cchmc.org/; Division of Biomedical Informatics, Children's Hospital Research Foundation, Cincinnati, OH, USA) [9], which predicts the relative solvent accessibility of amino acid residues and secondary structures in proteins using evolutionary profiles, the D816_I817delinsLL mutation was predicted to present physicochemical properties similar to those of D816V when compared with both the wild-type and the D816V mutation (Fig. 1D). In the physicochemical property prediction, both D816V and D816_I817delinsLL were predicted to be hydrophobic, whereas the wild type D816 and I817 were predicted to be negatively charged and hydrophobic, respectively. Considering these in-silico prediction results, we concluded that the clinical significance of D816_I817delinsLL mutation would be similar to that of the pathogenic D816V mutation.
In the follow-up BM examination, one month after the induction chemotherapy using idarubicin and cytarabine, the BM aspirate smear showed 14% of myeloblasts and the response was determined as resistant disease [10]. In the next follow-up BM examination, one month after re-induction chemotherapy using daunorubicin and cytarabine, 3% of myeloblasts were observed on the BM aspirate smear, and the patient was diagnosed with a morphological leukemia-free state.
This is the first report on a novel KIT INDEL mutation in exon 17, D816_I817delinsLL.
Acknowledgments
This study was supported by the Leading Foreign Research Institute Recruitment Program (No. 2011-0030034) through the NRF funded by the Ministry of Education, Science and Technology (MEST), a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (No. 2013-1320070) and grants from the Environmental Health Center funded by the Ministry of Environment, Republic of Korea.
References
1. Lovly C, Sosman J, Pao W. KIT. My cancer genome. Updated on Jun 16, 2015. http://www.mycancergenome.org/content/disease/acute-myeloid-leukemia/kit/?tab=0.
2. Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012; 366:1079–1089. PMID: 22417203.

3. Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015; 373:1136–1152. PMID: 26376137.

4. Wheeler S, Seegmiller A, Vnencak-Jones C. KIT c.2447A>T (D816V) mutation in acute myeloid leukemia. Updated on Sep 9, 2013. http://www.mycancergenome.org/content/disease/acute-myeloid-leukemia/kit/275/.
5. Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB, et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood. 2006; 107:3463–3468. PMID: 16384925.

6. National Comprehensive Cancer Network. Acute Myeloid Leukemia (Version 1.2015). Updated on Dec 3, 2014. http://www.nccn.org/professionals/physician_gls/pdf/aml.pdf.
7. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009; 4:1073–1081. PMID: 19561590.

8. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–249. PMID: 20354512.

9. Adamczak R, Porollo A, Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004; 56:753–767. PMID: 15281128.

10. Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, Estey EH, et al. Revised recommendations of the international working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003; 21:4642–4649. PMID: 14673054.

Fig. 1
(A) Bone marrow aspirate smear showing blasts presenting an abundant granular cytoplasm with perinuclear clearing, large orange-pink granules, and distinct Auer rods (Wright-Giemsa stain, ×1,000). (B) Result of real-time allele-specific PCR for KIT mutation (Real-Q C-KIT). The amplification curve of the patient sample (threshold cycle [Ct]: 29.4) and the amplification curve of positive control (Ct: 26.1) for KIT mutation screening are presented as the green and red line, respectively. The other amplification curves for KIT mutation genotyping such as D816V, D816Y, and D816H are not shown (not amplified). Rn is the normalized fluorescence signal of the reporter dye and ΔRn is Rn minus the baseline. The threshold of ΔRn is 0.1 and the cutoff value for KIT mutation positive is less than or equal to 38 as Ct. (C) Results of sequencing analysis of codon 815–817 of the KIT. The original sequence of the patient is presented (upper sequence) as well as the sequence of the wild-type allele after allele separation (middle sequence) and the sequence of the mutant allele after allele-separation (lower sequence). (D) Results of the in-silico prediction of protein structure and characteristics by SABLE. Wild-type (upper structure); D816V mutation (middle structure); D816_I817delinsLL mutation (lower structure). The highlighted background (blue) represents hydrophobic amino acids. The detail properties are explained in the POLYVIEW-2D documentation (http://polyview.cchmc.org/polyview_doc.html).
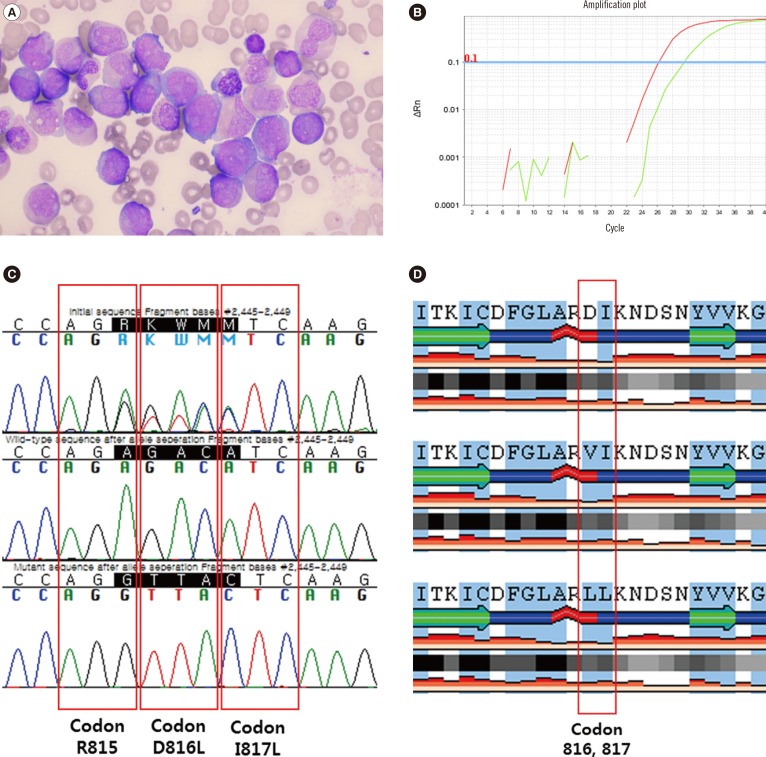
Table 1
Results of in-silico predictions about mutations in codon 816 and 817 of the KIT
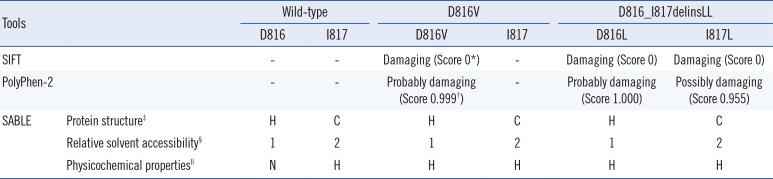
*SIFT score: the amino acid substitution is predicted to be damaging when the score is ≤0.05, and tolerated when the score is >0.05; †PolyPhen-2 score is the probability of the substitution being damaging; ‡SABLE Protein structure: H (helix), E (beta strand), C (coil); §SABLE Relative solvent accessibility: level of exposure (0-completely buried, 9-fully exposed); ∥SABLE Physicochemical properties: H-hydrophobic, N-negatively charged.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download