Abstract
Roseomonas is a genus of pink-pigmented nonfermentative bacilli. These slow-growing, gram-negative cocobacilli form pink-colored colonies on sheep blood agar. They differ from other pink-pigmented nonfermenters, including Methylobacterium, in morphology, biochemical characteristics, and DNA sequence. Roseomonas strains are rarely isolated in clinical laboratories; therefore, we report two cases in order to improve our ability to identify these pathogens. We isolated two strains of Roseomonas mucosa from the venous blood cultures of two patients, an 84-yr-old woman with common bile duct obstruction and a 17-yr-old male with acute myeloid leukemia who had an indwelling central-venous catheter for chemotherapy. The isolated strains were confirmed as R. mucosa by 16S rRNA sequencing.
The genus Roseomonas was initially identified as 42 strains of pink-pigmented gram-negative bacteria. From these strains, six genomospecies, all of which were considered Roseomonas [1], were proposed. Roseomonas constituted three named genomospecies, i.e., Roseomonas gilardii, R. cervicalis, and R. fauriae, and three unnamed genomospecies [2]. From 1992 to 2002, 36 strains of Roseomonas were isolated from blood cultures at the MD Anderson Cancer Center, and a new species, R. mucosa, was proposed on the basis of 16S rRNA sequencing and phylogenetic analysis [3]. Currently, the genus Roseomonas constitutes 15 valid species, including R. aquatica, R. aerilata, R. cervicalis, R. gilardii, R. lacus, R. mucosa, R. terrae, R. stagni, R. vinacea, R. fauriae, and other unnamed genomospecies.
Roseomonas are slow-growing, gram-negative bacteria. The colonies they form on sheep blood agar (SBA) are slightly fastidious, very mucoid, and almost runny. Microscopically, Roseomonas are nonvacuolated, plump coccobacilli that form mostly pairs and short chains. Although Roseomonas show low human pathogenicity, they can cause systemic infection in patients with underlying diseases or those who are immunocompromised [14567]. We isolated R. mucosa from the venous blood of two patients. We present the findings of these two cases and review the literature of R. mucosa bacteremia.
An 84-yr-old woman was admitted with jaundice and abdominal pain for three days. She had a history of cholecystitis and cholangitis and underwent cholecystectomy and percutaneous transhepatic biliary drainage. At admission, the values of AST, ALT, total bilirubin, and direct bilirubin were 73 IU/L, 44 IU/L, 14.93 mg/dL, and 13.5 mg/dL, respectively. Three sets of blood cultures were obtained before administration of antibiotics. Common bile duct obstruction was diagnosed by abdominal ultrasound and endoscopic retrograde cholangiopancreatography. After three-day incubation of blood culture bottles, positive signals were observed, and light pink-colored colonies formed on blood agar that was subcultured from one of the bottles (Fig. 1A). The isolate was identified as R. gilardii by the Vitek2 system (bioMerieux, Marcy l'Etoile, France). Matrix assisted laser desorption ionization – time of flight mass spectrometry (MALDI-TOF) analysis by Vitek MS (bioMerieux) failed because of the incomplete database, but 16S rRNA gene sequencing confirmed R. mucosa. After three days of cefotaxime and metronidazole combination therapy, the patient showed no signs of infection, including fever and chilling sensation. She was eventually discharged with no complications.
A 17-yr-old man was admitted to our hospital for consolidation chemotherapy. He had been diagnosed as having acute myeloid leukemia and received induction chemotherapy with no complications. During the consolidation chemotherapy, the patient exhibited fever and elevated C-reactive protein level with no focal infection. Ceftazidime and aminoglycoside combination as empirical antibiotics were initiated. Despite antibiotic therapy, fever persisted, and the C-reactive protein level further increased. Central catheter-related infection was suspected, and vancomycin was added to the antibiotic therapy after collection of two sets of central blood samples and three sets of venous blood samples. We found an isolate that was later identified as R. gilardii by the Vitek2 system but R. mucosa by 16S rRNA sequencing. Therefore, we reported this isolate as R. mucosa. Vancomycin and carbapenem combination therapy was continued, even though the patient did not exhibit significant clinical signs. The patient was discharged with no complications caused by the infection.
Roseomonas is found easily in the environment, including the soil, water, and air. The mechanism of infection caused by Roseomonas and its clinical significance are not well understood. Studies have proposed that Roseomonas can be found in turbid water from dirty or contaminated faucets [89]. However, although Roseomonas species have been isolated from specimens, only approximately 60% of isolates are clinically significant because of their low pathogenicity [4]. After their initial identification, several case reports have described clinically significant infections due to Roseomonas spp. Because of its low pathogenicity, Roseomonas can cause infection in patients with underlying disease or immunosuppression. Several cases have described catheter-related bacteremia with immunosuppression [5101112], bacteremia during chemotherapy for leukemia [1314], and peritonitis during continuous ambulatory peritoneal dialysis with end-stage renal disease [815]. However, R. mucosa infection in immunocompetent individuals has also been reported [6161718].
In our cases, R. mucosa was isolated by using conventional, routine methods. We used BACTEC Plus Aerobic/F and BACTEC Lytic/10 Anaerobic/F culture bottles and the BD Diagnostic System for blood culture. If a bottle showed a positive signal, subcultures were placed on SBA and MacConkey agar plates, which were incubated at 35℃ in a 5% CO2 incubator. Finally, the colonies were identified by using the Vitek2 system. Biochemical characterization of the isolated strains from our two patients showed positivity for catalase and urease, negativity for methanol oxidation, and a lack of UV light absorption [38], as summarized in Table 1. In order to identify prominent colonies of Rosemonas species, we attempted to subculture the isolates on potato cornmeal Tween 80 agar, which our institution uses for fungal subculture [19]; we found noticeable pink-colored runny mucoid colonies (Fig. 1B).
For molecular confirmation of the isolated strains, we performed 16S rRNA gene sequence analysis. The 16S rRNA gene was amplified in order to obtain a complete sequence, and the obtained sequence was then compared with the consensus sequence provided by GenBank (National Center for Biotechnology Information [NCBI]). The sequences were compared by using the Basic Local Alignment Search Tool provided by NCBI, and the result showed that the 16S rRNA gene sequence of R. mucosa with the highest similarity to the GenBank source matched with GenBank accession number NR_028857.1. The isolated Roseomonas species were susceptible to aminoglycosides (e.g., amikacin), carbapenems (e.g., imipenem), and fluoroquinolones (e.g., ciprofloxacin). The isolates had intermediate susceptibility to cefotaxime but were resistant to ampicillin and ceftazidime. This overall susceptibility pattern was consistent with the previous findings [237].
In summary, we report two cases of bacteremia due to R. mucosa, which is not exactly identified by conventional method of bacterial identification but confirmed by 16S rRNA gene sequence analysis.
References
1. Rihs JD, Brenner DJ, Weaver RE, Steigerwalt AG, Hollis DG, Yu VL. Roseomonas, a new genus associated with bacteremia and other human infections. J Clin Microbiol. 1993; 31:3275–3283. PMID: 8308122.
2. Dé I, Rolston KV, Han XY. Clinical significance of Roseomonas species isolated from catheter and blood samples: analysis of 36 cases in patients with cancer. Clin Infect Dis. 2004; 38:1579–1584. PMID: 15156446.
3. Han XY, Pham AS, Tarrand JJ, Rolston KV, Helsel LO, Levett PN. Bacteriologic characterization of 36 strains of Roseomonas species and proposal of Roseomonas mucosa sp nov and Roseomonas gilardii subsp rosea subsp nov. Am J Clin Pathol. 2003; 120:256–264. PMID: 12931556.
4. Struthers M, Wong J, Janda JM. An initial appraisal of the clinical significance of Roseomonas species associated with human infections. Clin Infect Dis. 1996; 23:729–733. PMID: 8909835.
5. Alcalá L, Vasallo FJ, Cercenado E, García-Garrote F, Rodríguez-Créixems M, Bouza E. Catheter-related bacteremia due to Roseomonas gilardii sp. nov. J Clin Microbiol. 1997; 35:2712. PMID: 9316945.
6. Fanella S, Schantz D, Karlowsky J, Rubinstein E. Septic arthritis due to Roseomonas gilardii in an immunocompetent adolescent. J Med Microbiol. 2009; 58:1514–1516. PMID: 19574413.
7. Wang CM, Lai CC, Tan CK, Huang YC, Chung KP, Lee MR, et al. Clinical characteristics of infections caused by Roseomonas species and antimicrobial susceptibilities of the isolates. Diagn Microbiol Infect Dis. 2012; 72:199–203. PMID: 22209515.
8. Sandoe JA, Malnick H, Loudon KW. A case of peritonitis caused by Roseomonas gilardii in a patient undergoing continuous ambulatory peritoneal dialysis. J Clin Microbiol. 1997; 35:2150–2152. PMID: 9230401.
9. Bibashi E, Sofianou D, Kontopoulou K, Mitsopoulos E, Kokolina E. Peritonitis due to Roseomonas fauriae in a patient undergoing continuous ambulatory peritoneal dialysis. J Clin Microbiol. 2000; 38:456–457. PMID: 10618142.
10. McLean TW, Rouster-Stevens K, Woods CR, Shetty AK. Catheter-related bacteremia due to Roseomonas species in pediatric hematology/oncology patients. Pediatr Blood Cancer. 2006; 46:514–516. PMID: 15782406.
11. Elshibly S, Xu J, McClurg RB, Rooney PJ, Millar BC, Alexander HD, et al. Central line-related bacteremia due to Roseomonas mucosa in a patient with diffuse large B-cell non-Hodgkins' lymphoma. Leuk Lymphoma. 2005; 46:611–614. PMID: 16019492.
12. Christakis GB, Perlorentzou S, Alexaki P, Megalakaki A, Zarkadis IK. Central line-related bacteraemia due to Roseomonas mucosa in a neutropenic patient with acute myeloid leukaemia in Piraeus, Greece. J Med Microbiol. 2006; 55:1153–1156. PMID: 16849738.
13. Salar A, Carratalà J, Zurita A, Gonzàlez-Barca E, Grañena A. Bacteremia caused by CDC group IV c-2 in a patient with acute leukemia. Haematologica. 1998; 83:670–672. PMID: 9718879.
14. Subudhi CP, Adedeji A, Kaufmann ME, Lucas GS, Kerr JR. Fatal Roseomonas gilardii bacteremia in a patient with refractory blast crisis of chronic myeloid leukemia. Clin Microbiol Infect. 2001; 7:573–575. PMID: 11683803.
15. Tsai SF, Chen CH, Shu KH, Wu MJ. Peritonitis caused by Roseomonas in a patient undergoing automated peritoneal dialysis: case report and literature review. Intern Med. 2012; 51:1721–1724. PMID: 22790133.
16. Ece G, Ruksen M, Akay A. Case report: cranioplasty infection due to gilardii at a university hospital in Turkey. Pan Afr Med J. 2013; 14:162. PMID: 23819004.

17. Nolan JS, Waites KB. Nosocomial ventriculitis due to Roseomonas gilardii complicating subarachnoid haemorrhage. J Infect. 2005; 50:244–251. PMID: 15780421.
18. Shokar NK, Shokar GS, Islam J, Cass AR. Roseomonas gilardii infection: case report and review. J Clin Microbiol. 2002; 40:4789–4791. PMID: 12454198.
19. Bard JD, Deville JG, Summanen PH, Lewinski MA. Roseomonas mucosa isolated from bloodstream of pediatric patient. J Clin Microbiol. 2010; 48:3027–3029. PMID: 20534804.
Fig. 1
(A) Roseomonas on blood agar showing nonhemolytic pale pink colonies after 72 hr incubation. (B) Roseomonas on potato cornmeal Tween 80 agar showing nonhemolytic, pink-colored, runny mucoid colonies after 24 hr incubation.
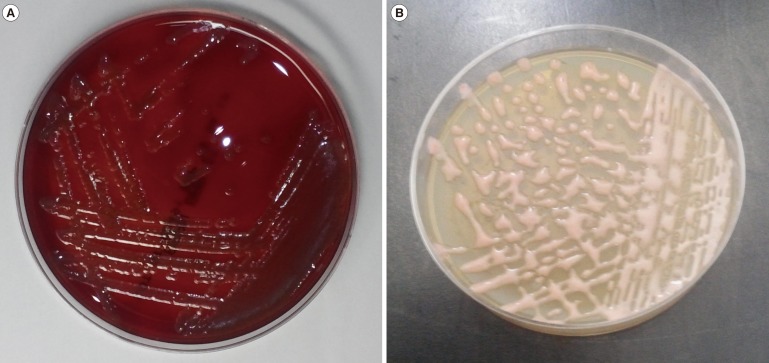
Table 1
Biochemical profiles of the isolated R. mucosa strains and the number of positive biochemical reactions of the reference strain
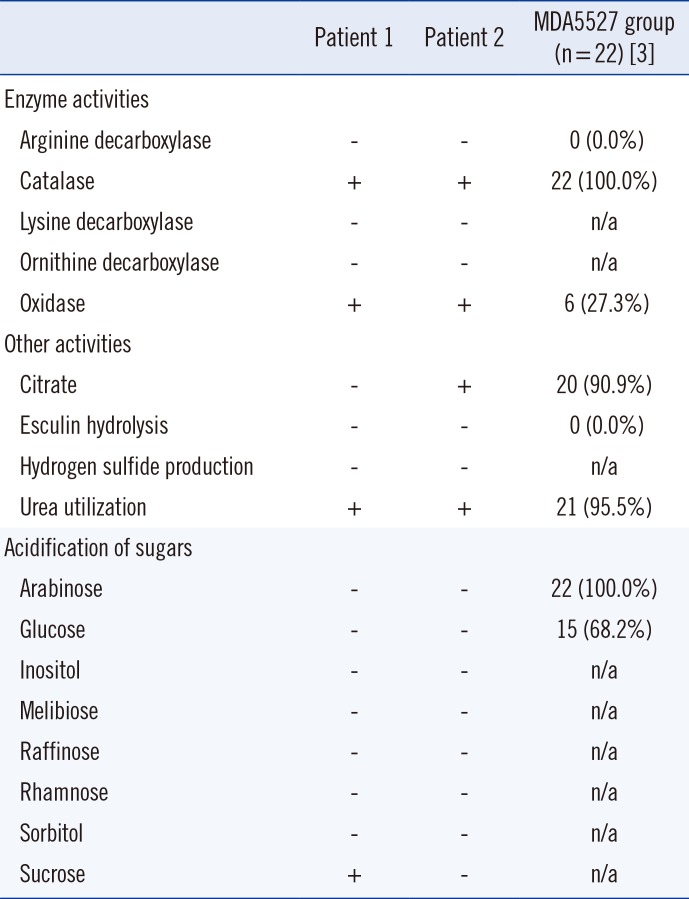
| Patient 1 | Patient 2 | MDA5527 group (n = 22) [3] | |
|---|---|---|---|
| Enzyme activities | |||
| Arginine decarboxylase | - | - | 0 (0.0%) |
| Catalase | + | + | 22 (100.0%) |
| Lysine decarboxylase | - | - | n/a |
| Ornithine decarboxylase | - | - | n/a |
| Oxidase | + | + | 6 (27.3%) |
| Other activities | |||
| Citrate | - | + | 20 (90.9%) |
| Esculin hydrolysis | - | - | 0 (0.0%) |
| Hydrogen sulfide production | - | - | n/a |
| Urea utilization | + | + | 21 (95.5%) |
| Acidification of sugars | |||
| Arabinose | - | - | 22 (100.0%) |
| Glucose | - | - | 15 (68.2%) |
| Inositol | - | - | n/a |
| Melibiose | - | - | n/a |
| Raffinose | - | - | n/a |
| Rhamnose | - | - | n/a |
| Sorbitol | - | - | n/a |
| Sucrose | + | - | n/a |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download