Dear Editor,
Clostridium tertium is an endospore-forming anaerobic gram–positive bacillus. This organism is aerotolerant and is easily decolorized in Gram-stained smears, often leading to mistaken identification as a gram-negative organism. The major risk factors for C. tertium bacteremia are hematological disease, intestinal mucosal injury, and history of exposure to β-lactam antibiotics (such as the third and fourth generation cephalosporins) where fever and leukopenia are often seen in patients [1]. We report two cases of C. tertium isolated from blood culture, one of which was successfully identified by using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis and subsequent 16S ribosomal DNA sequencing.
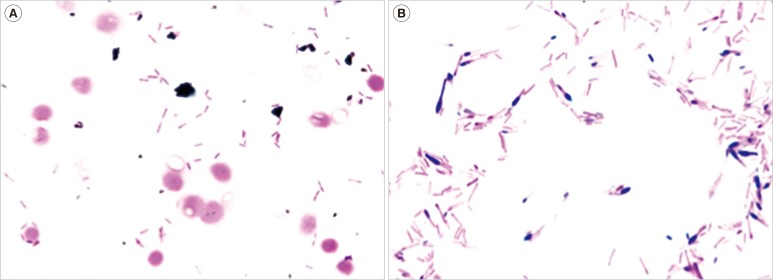
Case 1. A man in his 50s collapsed at home, became hypothermic with shock, and was hospitalized. On day 2 after hospitalization, two sets of blood cultures were taken, and antimicrobial treatment began with tazobactam/piperacillin (TAZ/PIPC) (2.25g×4/day). On day 3, he died and an aerobic isolate from the blood cultures became positive (BacT/Alert3D; bioMérieux, Tokyo, Japan) where gram-negative bacilli were recovered and sub-cultured aerobically at 35℃ for 24 hr (Fig. 1A). Shiny colonies (about 1 mm in diameter) grew on blood agar plates, but not on BTB agar plates. Further, anaerobical culture of blood on Brucella HK agar plates at 35℃ for 48 hr yielded gray colonies (about 3 mm in diameter). Gram stain showed spore-forming gram-variable bacilli (Fig. 1B). We therefore suspected Clostridium species. The organism was identified as C. tertium on the basis of the results of an automated identification apparatus (VITEK2 Compact; Sysmex bioMérieux), an identification kit (Rap ID ANAII; Amuko, Tokyo, Japan), and negative catalase test results. Later examination by MALDI-TOF MS-Biotyper (Bruker Daltonics, Yokohama, Japan) confirmed the organism as C. tertium with an identification log score 2.030 (2.000-2.299: secure genus identification and probable species identification). Antibiotic susceptibility showed that the minimum inhibitory concentration (MIC) value against C. tertium was 8 µg/mL for TAZ/PIPC, 16 µg/mL for cefotaxime (CTX), and 1 µg/mL for metronidazole.
Case 2. A man in his 60s had been diagnosed as having acute myelogenous leukemia and hospitalized for chemotherapy. Two days after beginning chemotherapy, he presented with fever (38.0℃) and his leukocyte count declined to 1.3×109/L. We began an antimicrobial treatment with cefepime (CFPM) (1g ×4/day) which proved ineffective. Three blood cultures gave negative results. On day 14 after the start of the chemotherapy, two sets of blood cultures were taken; and the antimicrobial therapy was changed to meropenem (MEPM) (1g ×3/day). On day 16, one of the two sets of aerobic blood cultures grew Gram-variable bacilli similar to Case 1, leading us again to suspect a Clostridium species. Antibiotic susceptibility tests showed MICs for CTX at ≥32 µg/mL, ceftriaxone at ≥32 µg/mL, cefozopran at 16 µg/mL, MEPM at ≤0.25 µg/mL, and vancomycin (VCM) at ≤1 µg/mL. The antimicrobial treatment regimen was then supplemented with VCM (1g ×3/day). Blood was aerobically cultured and colonies similar to Case 1 grew on blood, chocolate, and Brucella HK agar plates but not on BTB agar plates. VITEK2 compact and Rap ID ANA II analyses reported the presence of the test strain as C. clostridioforme; however, we doubted that report for several reasons. C. clostridioforme is anaerobic, so it can be mistaken for a gram-negative bacillus similar to C. tertium. Further, C. clostridioforme is usually suceptible to β-lactam antibiotics. The clinical strain formed endospores in anaerobic culture while C. clostridioforme does not form endospores but rather a so-called 'football-form' in Gram stains [1].
Consequently, we performed further identification using MALDI-TOF MS-Biotyper (Bruker Daltonics). That analysis reported the clinical strain as C. tertium with an identification log score of 2.030. Sequencing of the 16S rDNA gene showed complete identity to C. tertium. C. tertium was considered a pathogenic bacterium in the case because it had been isolated from blood culture and because of the neutropenia. C. tertium is resistant to the third and fourth generation cephalosporins and aminoglycosides, and VCM and/or imipenem are often used therapeutically to treat C. tertium infection [23]. MEPM and VCM were administered for seven days and the patient survived.
Given the difficulties in differentiating between the several Clostridium species, our work underscores the necessity of using a variety of techniques to assist clinical identification. Our study suggests identification of bacterial species by combining MALDI-TOF MS with 16S rRNA gene sequencing provides a reliable means to obtain rapid identification of pathogens in the clinical setting, removing the ambiguity around poorly differentiable strains such as those of Clostridium species.
Acknowledgments
The authors thank Jim Nelson for his editorial assistance. This study was supported by institutional grant for RCNID.
Go to : 
Notes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
Go to : 
References
1. Miller DL, Brazer S, Murdoch D, Reller LB, Corey GR. Significance of Clostridium tertium bacteremia in neutropenic and nonneutropenic patients: review of 32 cases. Clin Infect Dis. 2001; 32:975–978. PMID: 11247721.
2. Steyaert S, Peleman R, Vaneechoutte M, De Baere T, Claeys G, Verschraegen G. Septicemia in neutropenic patients infected with Clostridium tertium resistant to cefepime and other expanded-spectrum cephalosporins. J Clin Microbiol. 1999; 37:3778–3779. PMID: 10523601.
3. Speirs G, Warren RE, Rampling A. Clostridium tertium septicemia in patients with neutropenia. J Infect Dis. 1988; 158:1336–1340. PMID: 3198941.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download