Staphylococcus aureus is a significant pathogen in both nosocomial and community infections. This bacterium is capable of colonizing various bodily sites, especially the nose [
1]. Nasal carriage of
S. aureus or methicillin-resistant
S. aureus (MRSA) is considered a risk factor for further endogenous infections such as sepsis [
2]. Various carriage rates of nasal
S. aureus have been reported, and recent data suggestsa carriage rate of 20-30%. Moreover, scientific interest has focused on nasal community-associated MRSA (CA-MRSA), with respect to nasal prevalence, antibiotic resistance, and molecular characterization [
34]. MRSA strains first emerged in hospitals in the early 1960s, and several clones have become prevalent in healthcare settings worldwide. However, several community infection-associated clones, referred to as CA-MRSA, emerged in the 1990s and changed the concept of MRSA. Worse still, CA-MRSA tend to be multi-drug resistant and to invade hospital settings. Nasal carriers of CA-MRSA strains may play a role in infection transmission between community and hospital settings. Therefore, it is of great importance to detect nasal carriage of
S. aureus among inpatients at hospital admission.
The healthcare-associated MRSA (HA-MRSA) sequence type (ST) 239 clone is prevalent in China, whereas methicillin-sensitive
S. aureus (MSSA) strains show great diversity in molecular characteristics [
5]. Several CA-MRSA clones have been reported from China, and ST59, or its single locus variant ST338, is the most common clone [
56]. Nevertheless, molecular epidemiology of nasal
S. aureus among healthy individuals and community populations from China indicate molecular heterogeneity.Limited data on nasal
S. aureus among inpatients was reported. In the present study, we analyzed nasal carriage rates of
S. aureus among newlyadmitted inpatients by microbiologic examination, antimicrobial susceptibility testing, and molecular characterization.
A total of 66 nasal
S. aureus isolates were collected from 292 patients with in 48 hr safter admission to a tertiary hospital in Shanghai, China. Bacterial identification was performed by a combination of phenotypic tests, such as microscopic examination andproduction of coagulase and DNase [
78]. Molecular identification was verified by amplification of the
nuc gene with specific primers [
8]. Susceptibility testing was carried out by using the disk diffusion method according to the CLSI guidelines [
9]. Tested antimicrobials included penicillin (10 units), cefoxitin (30 µg), teicoplanin (30 µg), gentamicin (10 µg), kanamycin (30 µg), tobramycin (10 µg), erythromycin (15 µg), clindamycin (2 µg), quinupristin/dalfopristin (15 µg), tetracycline (30 µg), minocycline (30 µg), ciprofloxacin (5 µg), sulfamethoxazole/trimethoprim (25 µg), chloramphenicol (30 µg), rifampicin (5 µg), linezolid (30 µg), and fusidic acid (10 µg). Susceptibility to penicillin was also observed by the penicillin zone-edge test. Inducible resistance to clindamycin was tested by using the D-test, and high-level mupirocin resistance (MUP
Hi) was screened by using a mupirocin disk (200 µg).
S. aureus ATCC 25923 was used as the control strain for susceptibility testing.
MRSA strains were screened by cefoxitin disk and confirmed by detection of the
mecA gene. The
erm genes (
ermA,
ermB, and
ermC) and
ileS-2 gene were detected by PCR as previously described [
1011]. Multi-locus sequence typing (MLST) and spa typing, with additional SCC
mec typing for MRSA strains, were performed [
101112]. Analysis of these molecular characteristics was conducted by grouping isolates with eBURST based on ST. Moreover, a series of clinically significant toxin genes encoding staphylococcal enterotoxins (
sea-see and
seg-sej), exfoliative toxin (
eta and
etb), toxic shock syndrome toxin 1 (
tst), and Panton-Valentine leukocidin (
pvl) were detected [
13].
By cefoxitin screening and
mecA gene detection, 18 out of 66 (27.3%) nasal
S. aureus isolates were
mecA-positive MRSA. The results of antimicrobial susceptibility testing revealed that all isolates were sensitive to linezolid, teicoplanin, and quinupristin/dalfopristin. Two isolates, one MRSA strain and one MSSA strain isolated from the same patient, exhibited the MUP
Hi phenotype and harbored the
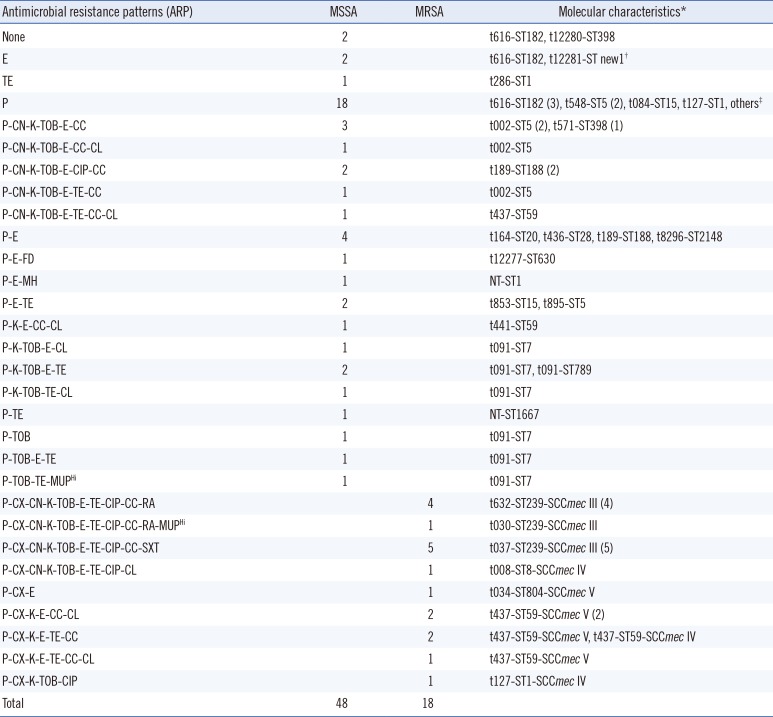
ileS-2 gene. High resistance ratesfor penicillin (92.4%), erythromycin (60.6%), kanamycin (45.5%), tobramycin (40.9%), tetracycline (37.9%), clindamycin (36.4%), and gentamicin (28.8%) were detected. Resistance rates for ciprofloxacin, chloramphenicol, sulfamethoxazole/trimethoprim, rifampicin, minocycline, and fusidic acid were 21.2%, 13.6%, 7.6%, 7.6%, 1.5%, and 1.5%, respectively.The antimicrobial resistance patterns (ARP) of the nasal
S. aureus isolates, including 18 MRSA and 48 MSSA are shown in
Table 1.
By molecular typing, 10 out of 18 (55.6%) MRSA isolates were HA-MRSA clones, all of which were ST239-SCC
mec III with different spa-types. The remaining eight MRSA isolates were genotypic CA-MRSA strains, a half of which were t437-ST59-SCC
mec V. On the other hand, MSSA strains displayed diverse molecular characteristics with some prominent clones, including t091-ST7 (six isolates), t616-ST182 (five isolates), t189-ST188 (four isolates), and t002-ST5 (four isolates). Toxin gene profiling revealed that most isolates (35/66, 53%) did not harbor any toxin genes. HA-MRSA strains tended to carry the sea gene, while CA-MRSA strains usually carried the
seb gene. Moreover, all four CA-MRSA isolates, which were t437-ST59-SCC
mec V, were positive for the
pvl gene. Different staphylococcal enterotoxin genes were found in some MSSA isolates, especially ST5 and ST182 clones. Detailed molecular characterization and toxin gene profilesare shown in
Table 1 and the Supplemental Data
Table S1.
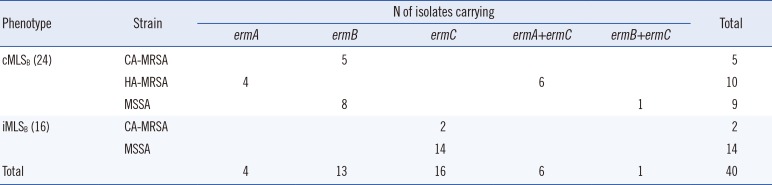
According to our D-test results, the phenotypes cMLSB, characterized by resistance to both erythromycin and clindamycin, and iMLSB (positive D-test) were detected in 24 and 16 isolates, respectively. However, no isolates exhibited the MSB phenotype, characterized by susceptibility to clindamycin and erythromycin resistance with a negative D-test.
The
erm genes were detected, and their distribution among different strains, including CA-MRSA, HA-MRSA, and MSSA, was explored. As shown in
Table 2, all 10 HA-MRSA strains displayed the cMLS
B phenotype, which was mediated by the
ermA or
ermA/ermC genes. CA-MRSA and MSSA strains exhibiting the cMLS
B phenoty pecarried the
ermB gene. All D-test positive strains, both MSSA and CA-MRSA, were only positive for the
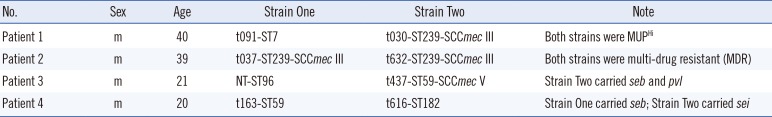
ermC gene. Unexpectedly, four patients were found to be co-colonized with two nasal
S. aureus strains with different molecular characteristics. Brief information on these four patients and the strains is listed in
Table 3.
This study investigated the status of nasal S.aureus from newlyadmitted inpatients, and the results revealed that 62 out of 292 (20.5%) inpatients carried S. aureus in the nose. Among the 62 inpatients, 66 S. aureus isolates were detected, as four patients were co-colonized with two nasal S. aureus strains with different molecular characteristics. Of these 66 S. aureus isolates, 18 were MRSA strains including 10 HA-MRSA and 8 CA-MRSA strains. High resistance rates for penicillin and erythromycin were observed for both MSSA and MRSA strains.
Nasal colonization of
S. aureus can be seen as a risk factor for further endogenous infection [
2]. In this study, 20.5% of newlyadmitted inpatients carried
S. aureus in the nose, suggesting that it is essential to detect and eliminatenasal
S. aureus in inpatients at hospital admission, especially those of high-risk departments, such as orthopedics or the burn unit [
1415]. The antibiotic erythromycin was commonly used for combating
S. aureus in China, yet high resistance to erythromycin was observed in these nasal isolates. In
Table 2, we show that different kinds of strains displayed diverse resistance phenotypesmediated by various resistance genes. It is difficult to eliminate or treat nasal
S. aureus efficiently; mupirocin is a good option despite the fact that some mupirocin-resistant
S. aureus strains were detected in the present study.
Molecular characterization of the nasal
S. aureus isolates revealed that all 10 HA-MRSA strains were ST239-SCC
mec III, a major clone causing nosocomial infection, and that four out of eight CA-MRSA strains were t437-ST59-SCC
mec V, reported to be a prevalent clone in community settings. As reported, HA-MRSA and CA-MRSA strains may co-exist with the interaction and dynamic of hospital and community populations [
16]. In the present study, molecular data typically provided microbiological and epidemiological informationon the role of patients in exchanging epidemic CA-MRSA and HA-MRSA strains. Therefore, the nasal carrier patients can be regarded as an important risk factor for the introduction of CA-MRSA into hospitals. It would also be helpful to prevent and monitor hospital infections if some measures were taken to detect the nasal carriage status of inpatientsin China.
Unexpectedly, it was found that four patients were co-colonized by two nasal S. aureus strains. It was suspected when distinct bacterial colonies with different morphology and hemolysis were observed on blood agar plates, and was confirmed by molecular typing.In one patient, both strains (one MSSA t091-ST7 and one MRSA t030-ST239-SCCmec III) exhibited the MUPHi phenotype and harbored the ileS-2 gene, indicating potential horizontal resistance determinant transfer in vivo in the nasal cavity.
In this study, antimicrobial resistance and molecular characteristics of nasal S. aureus isolates from newlyadmitted inpatients were investigated. All isolates were sensitive to linezolid, teicoplanin, and quinupristin/dalfopristin, but high resistance against erythromycin was detected. Molecular evidence suggests that patients play an indispensable role in transmitting epidemic CA-MRSA and HA-MRSA strains in different environments, thus making it essential to detect the nasal carriage status of inpatients to control emerging genotypic CA-MRSA strains in hospitals.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download