1. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006; 12:826–836. PMID:
16882287.

2. Sevillano E, Valderrey C, Canduela MJ, Umaran A, Calvo F, Gallego L. Resistance to antibiotics in clinical isolates of Pseudomonas aeruginosa. Pathol Biol (Paris). 2006; 54:493–497. PMID:
17027190.
3. Oh EJ, Lee S, Park YJ, Park JJ, Park K, Kim SI, et al. Prevalence of metallo-beta-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-beta-lactamase. J Microbiol Methods. 2003; 54:411–418. PMID:
12842488.
4. Jeong SH, Bae IK, Park KO, An YJ, Sohn SG, Jang SJ, et al. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J Microbiol. 2006; 44:423–431. PMID:
16953178.
5. Lee K, Park AJ, Kim MY, Lee HJ, Cho JH, Kang JO, et al. Metallo-beta-lactamase-producing Pseudomonas spp. in Korea: high prevalence of isolates with VIM-2 type and emergence of isolates with IMP-1 type. Yonsei Med J. 2009; 50:335–339. PMID:
19568593.
6. Yong D, Choi YS, Roh KH, Kim CK, Park YH, Yum JH, et al. Increasing prevalence and diversity of metallo-beta-lactamases in Pseudomonas spp., Acinetobacter spp., and Enterobacteriaceae from Korea. Antimicrob Agents Chemother. 2006; 50:1884–1886. PMID:
16641469.
7. Sung JY, Koo SH, Cho HH, Kwon KC. Dissemination of an AbaR-type resistance island in multidrug-resistant Acinetobacter baumannii global clone 2 in Daejeon of Korea. Ann Clin Microbiol. 2013; 16:75–80.
8. Chung H-S, Lee Y, Park ES, Lee DS, Ha EJ, Kim M, et al. Characterization of the multidrug-resistant Acinetobacter species causing a nosocomial outbreak at intensive care units in a Korean teaching hospital: suggesting the correlations with the clinical and environmental samples, including respiratory tract-related instruments. Ann Clin Microbiol. 2014; 17:29–34.
10. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007; 20:440–458. PMID:
17630334.
11. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001; 7:88–91. PMID:
11298149.
12. Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003; 41:4623–4629. PMID:
14532193.
13. Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011; 3:189–193. PMID:
22530087.
14. Markelz AE, Mende K, Murray CK, Yu X, Zera WC, Hospenthal DR, et al. Carbapenem susceptibility testing errors using three automated systems, disk diffusion, Etest, and broth microdilution and carbapenem resistance genes in isolates of Acinetobacter baumannii-calcoaceticus complex. Antimicrob Agents Chemother. 2011; 55:4707–4711. PMID:
21807971.

15. Kim HJ, Kim YJ, Yong DE, Lee K, Park JH, Lee JM, et al. Loop-mediated isothermal amplification of vanA gene enables a rapid and naked-eye detection of vancomycin-resistant enterococci infection. J Microbiol Methods. 2014; 104:61–66. PMID:
24925601.

16. Labarca JA, Salles MJ, Seas C, Guzmán-Blanco M. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit Rev Microbiol. 2014; 1–17. PMID:
25159043.
17. Kim BM, Jeon EJ, Jang JY, Chung JW, Park J, Choi JC, et al. Four year trend of carbapenem-resistance in newly opened ICUs of a university-affiliated hospital of South Korea. Tuberc Respir Dis (Seoul). 2012; 72:360–366. PMID:
23227077.

18. Tripathi A, Shukla SK, Singh A, Prasad KN. A new approach of real time polymerase chain reaction in detection of vancomycin-resistant enterococci and its comparison with other methods. Indian J Med Microbiol. 2013; 31:47–52. PMID:
23508429.

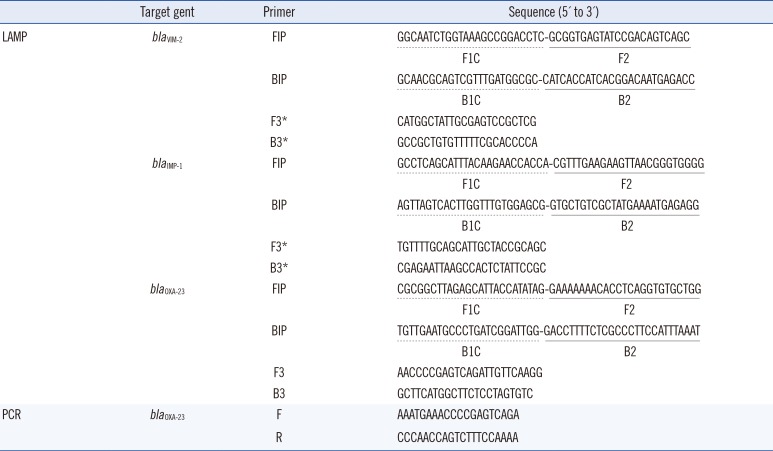
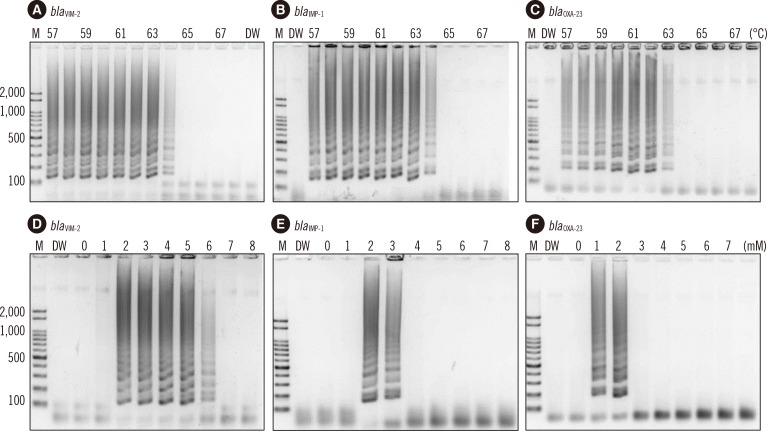
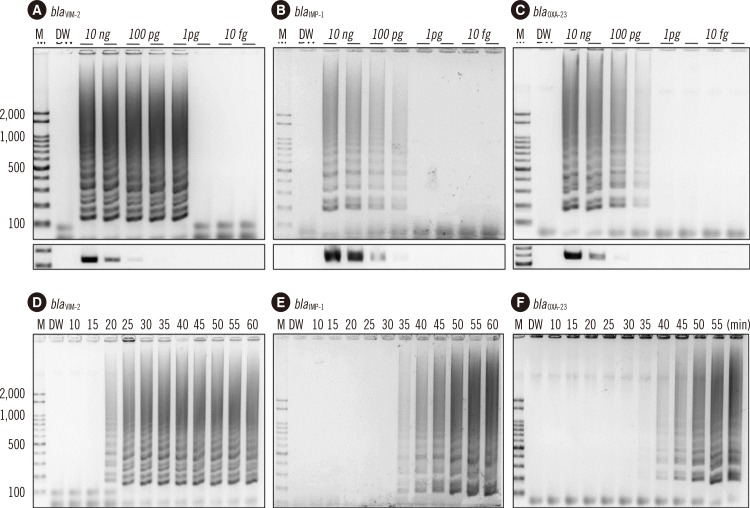
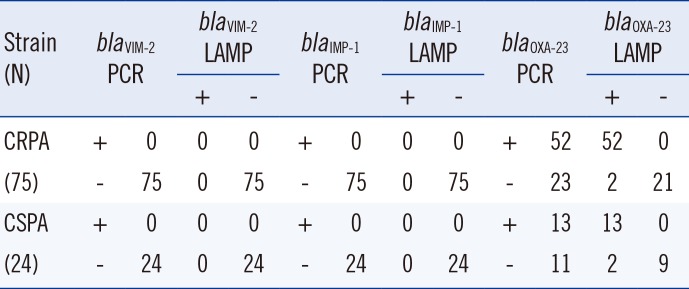
19. Kojima T, Shibata N, Ikedo M, Arakawa Y. Development and evaluation of novel loop-mediated isothermal amplification for rapid detection of bla(IMP-1) and bla(VIM-2) genes. Kansenshogaku Zasshi. 2006; 80:405–412. PMID:
16922484.
20. Lee Y, Kim C-K, Chung H-S, Yong D, Jeong SH, Lee K, et al. Increasing carbapenem-resistant gram-negative bacilli and decreasing metallo-β-lactamase producers over eight years from Korea. Yonsei Med J. 2015; 56:572–577. PMID:
25684011.

21. Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009; 53:4783–4788. PMID:
19738025.
22. Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2006; 50:1633–1641. PMID:
16641429.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download